{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# CNTK 204: Sequence to Sequence Networks with Text Data\n",
"\n",
"\n",
"## Introduction and Background\n",
"\n",
"This hands-on tutorial will take you through both the basics of sequence-to-sequence networks, and how to implement them in the Microsoft Cognitive Toolkit. In particular, we will implement a sequence-to-sequence model to perform grapheme to phoneme translation. We will start with some basic theory and then explain the data in more detail, and how you can download it.\n",
"\n",
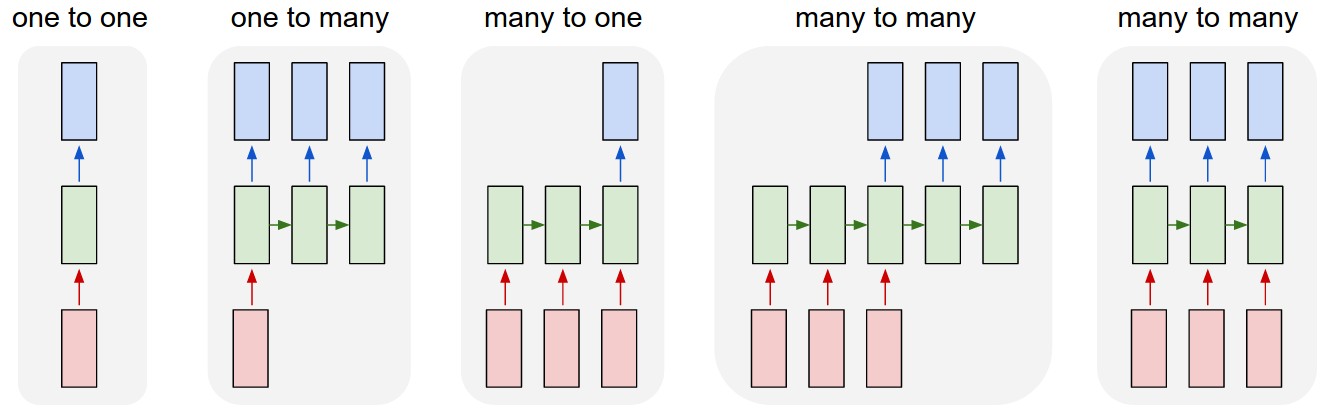
"Andrej Karpathy has a [nice visualization](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) of the five paradigms of neural network architectures:\n",
"\n",
" \n",
"\n",
"In this tutorial, we are going to be talking about the fourth paradigm: many-to-many, also known as sequence-to-sequence networks. The input is a sequence with a dynamic length, and the output is also a sequence with some dynamic length. It is the logical extension of the many-to-one paradigm in that previously we were predicting some category (which could easily be one of `V` words where `V` is an entire vocabulary) and now we want to predict a whole sequence of those categories.\n",
"\n",
"The applications of sequence-to-sequence networks are nearly limitless. It is a natural fit for machine translation (e.g. English input sequences, French output sequences); automatic text summarization (e.g. full document input sequence, summary output sequence); word to pronunciation models (e.g. character [grapheme] input sequence, pronunciation [phoneme] output sequence); and even parse tree generation (e.g. regular text input, flat parse tree output).\n",
"\n",
"## Basic theory\n",
"\n",
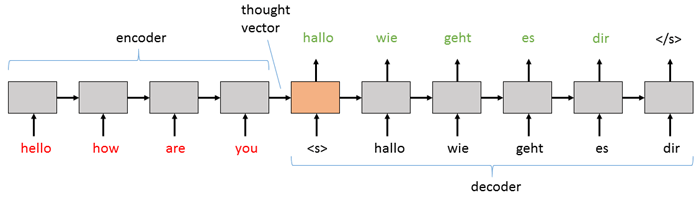
"A sequence-to-sequence model consists of two main pieces: (1) an encoder; and (2) a decoder. Both the encoder and the decoder are recurrent neural network (RNN) layers that can be implemented using a vanilla RNN, an LSTM, or GRU cells (here we will use LSTM). In the basic sequence-to-sequence model, the encoder processes the input sequence into a fixed representation that is fed into the decoder as a context. The decoder then uses some mechanism (discussed below) to decode the processed information into an output sequence. The decoder is a language model that is augmented with some \"strong context\" by the encoder, and so each symbol that it generates is fed back into the decoder for additional context (like a traditional LM). For an English to German translation task, the most basic setup might look something like this:\n",
"\n",
"
\n",
"\n",
"In this tutorial, we are going to be talking about the fourth paradigm: many-to-many, also known as sequence-to-sequence networks. The input is a sequence with a dynamic length, and the output is also a sequence with some dynamic length. It is the logical extension of the many-to-one paradigm in that previously we were predicting some category (which could easily be one of `V` words where `V` is an entire vocabulary) and now we want to predict a whole sequence of those categories.\n",
"\n",
"The applications of sequence-to-sequence networks are nearly limitless. It is a natural fit for machine translation (e.g. English input sequences, French output sequences); automatic text summarization (e.g. full document input sequence, summary output sequence); word to pronunciation models (e.g. character [grapheme] input sequence, pronunciation [phoneme] output sequence); and even parse tree generation (e.g. regular text input, flat parse tree output).\n",
"\n",
"## Basic theory\n",
"\n",
"A sequence-to-sequence model consists of two main pieces: (1) an encoder; and (2) a decoder. Both the encoder and the decoder are recurrent neural network (RNN) layers that can be implemented using a vanilla RNN, an LSTM, or GRU cells (here we will use LSTM). In the basic sequence-to-sequence model, the encoder processes the input sequence into a fixed representation that is fed into the decoder as a context. The decoder then uses some mechanism (discussed below) to decode the processed information into an output sequence. The decoder is a language model that is augmented with some \"strong context\" by the encoder, and so each symbol that it generates is fed back into the decoder for additional context (like a traditional LM). For an English to German translation task, the most basic setup might look something like this:\n",
"\n",
" \n",
"\n",
"The basic sequence-to-sequence network passes the information from the encoder to the decoder by initializing the decoder RNN with the final hidden state of the encoder as its initial hidden state. The input is then a \"sequence start\" tag (`
\n",
"\n",
"The basic sequence-to-sequence network passes the information from the encoder to the decoder by initializing the decoder RNN with the final hidden state of the encoder as its initial hidden state. The input is then a \"sequence start\" tag (`` in the diagram above) which primes the decoder to start generating an output sequence. Then, whatever word (or note or image, etc.) it generates at that step is fed in as the input for the next step. The decoder keeps generating outputs until it hits the special \"end sequence\" tag (`` above).\n",
"\n",
"A more complex and powerful version of the basic sequence-to-sequence network uses an attention model. While the above setup works well, it can start to break down when the input sequences get long. At each step, the hidden state `h` is getting updated with the most recent information, and therefore `h` might be getting \"diluted\" in information as it processes each token. Further, even with a relatively short sequence, the last token will always get the last say and therefore the thought vector will be somewhat biased/weighted towards that last word. To deal with this problem, we use an \"attention\" mechanism that allows the decoder to look not only at all of the hidden states from the input, but it also learns which hidden states, for each step in decoding, to put the most weight on. We will discuss an attention implementation in a later version of this tutorial."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Problem: Grapheme-to-Phoneme Conversion\n",
"\n",
"The [grapheme](https://en.wikipedia.org/wiki/Grapheme) to [phoneme](https://en.wikipedia.org/wiki/Phoneme) problem is a translation task that takes the letters of a word as the input sequence (the graphemes are the smallest units of a writing system) and outputs the corresponding phonemes; that is, the units of sound that make up a language. In other words, the system aims to generate an unambigious representation of how to pronounce a given input word.\n",
"\n",
"### Example\n",
"\n",
"The graphemes or the letters are translated into corresponding phonemes: \n",
"\n",
"> **Grapheme** : **|** T **|** A **|** N **|** G **|** E **|** R **|** \n",
"**Phonemes** : **|** ~T **|** ~AE **|** ~NG **|** ~ER **|** null **|** null **|** \n",
"\n",
"\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Task and Model Structure\n",
"\n",
"As discussed above, the task we are interested in solving is creating a model that takes some sequence as an input, and generates an output sequence based on the contents of the input. The model's job is to learn the mapping from the input sequence to the output sequence that it will generate. The job of the encoder is to come up with a good representation of the input that the decoder can use to generate a good output. For both the encoder and the decoder, the LSTM does a good job at this.\n",
"\n",
"We will use the LSTM implementation from the CNTK Blocks library. This implements the \"smarts\" of the LSTM and we can more or less think of it as a black box. What is important to understand, however, is that there are two pieces to think of when implementing an RNN: the recurrence, which is the unrolled network over a sequence, and the block, which is the piece of the network run for each element of the sequence. We only need to implement the recurrence.\n",
"\n",
"It helps to think of the recurrence as a function that keeps calling `step(x)` on the block (in our case, LSTM). At a high level, it looks like this:\n",
"\n",
"```\n",
"class LSTM {\n",
" float hidden_state\n",
"\n",
" init(initial_value):\n",
" hidden_state = initial_value\n",
"\n",
" step(x):\n",
" hidden_state = LSTM_function(x, hidden_state)\n",
" return hidden_state\n",
"}\n",
"```\n",
"\n",
"So, each call to the `step(x)` function takes some input `x`, modifies the internal `hidden_state`, and returns it. Therefore, with every input `x`, the value of the `hidden_state` evolves. Below we will import some required functionality, and then implement the recurrence that makes use of this mechanism."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Importing CNTK and other useful libraries\n",
"\n",
"CNTK is a Python module that contains several submodules like `io`, `learner`, `graph`, etc. We make extensive use of numpy as well."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"from __future__ import print_function\n",
"import numpy as np\n",
"import os\n",
"from cntk import Trainer, Axis\n",
"from cntk.io import MinibatchSource, CTFDeserializer, StreamDef, StreamDefs, INFINITELY_REPEAT\n",
"from cntk.learner import momentum_sgd, momentum_as_time_constant_schedule, learning_rate_schedule, UnitType\n",
"from cntk.ops import input_variable, cross_entropy_with_softmax, classification_error, sequence, past_value, future_value, element_select, \\\n",
" alias, hardmax, placeholder_variable, combine, parameter, plus, times\n",
"from cntk.ops.functions import CloneMethod\n",
"from cntk.blocks import LSTM, Stabilizer\n",
"from cntk.initializer import glorot_uniform\n",
"from cntk.utils import get_train_eval_criterion, get_train_loss\n",
"# Select the right target device when this notebook is being tested:\n",
"if 'TEST_DEVICE' in os.environ:\n",
" import cntk\n",
" if os.environ['TEST_DEVICE'] == 'cpu':\n",
" cntk.device.set_default_device(cntk.device.cpu())\n",
" else:\n",
" cntk.device.set_default_device(cntk.device.gpu(0))\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Downloading the data\n",
"\n",
"In this tutorial we will use a lightly pre-processed version of the CMUDict (version 0.7b) dataset from http://www.speech.cs.cmu.edu/cgi-bin/cmudict. The CMUDict data is the Carnegie Mellon University Pronouncing Dictionary is an open-source machine-readable pronunciation dictionary for North American English. The data is in the CNTKTextFormatReader format. Here is an example sequence pair from the data, where the input sequence (S0) is in the left column, and the output sequence (S1) is on the right:\n",
"\n",
"```\n",
"0\t|S0 3:1 |# \t|S1 3:1 |# \n",
"0\t|S0 4:1 |# A\t|S1 32:1 |# ~AH\n",
"0\t|S0 5:1 |# B\t|S1 36:1 |# ~B\n",
"0\t|S0 4:1 |# A\t|S1 31:1 |# ~AE\n",
"0\t|S0 7:1 |# D\t|S1 38:1 |# ~D\n",
"0\t|S0 12:1 |# I\t|S1 47:1 |# ~IY\n",
"0\t|S0 1:1 |# \t|S1 1:1 |# \n",
"```\n",
"\n",
"The code below will download the required files (training, the single sequence above for validation, and a small vocab file) and put them in a local folder (the training file is ~34 MB, testing is ~4MB, and the validation file and vocab file are both less than 1KB)."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"import requests\n",
"\n",
"def download(url, filename):\n",
" \"\"\" utility function to download a file \"\"\"\n",
" response = requests.get(url, stream=True)\n",
" with open(filename, \"wb\") as handle:\n",
" for data in response.iter_content():\n",
" handle.write(data)\n",
"\n",
"data_dir = os.path.join('..', 'Examples', 'SequenceToSequence', 'CMUDict', 'Data')\n",
"# If above directory does not exist, just use current.\n",
"if not os.path.exists(data_dir):\n",
" data_dir = '.'\n",
"\n",
"valid_file = os.path.join(data_dir, 'tiny.ctf')\n",
"train_file = os.path.join(data_dir, 'cmudict-0.7b.train-dev-20-21.ctf')\n",
"vocab_file = os.path.join(data_dir, 'cmudict-0.7b.mapping')\n",
"\n",
"files = [valid_file, train_file, vocab_file]\n",
"\n",
"for file in files:\n",
" if os.path.exists(file):\n",
" print(\"Reusing locally cached: \", file)\n",
" else:\n",
" url = \"https://github.com/Microsoft/CNTK/blob/v2.0.beta7.0/Examples/SequenceToSequence/CMUDict/Data/%s?raw=true\"%file\n",
" print(\"Starting download:\", file)\n",
" download(url, file)\n",
" print(\"Download completed\")\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Select the notebook run mode\n",
"\n",
"There are two run modes:\n",
"- *Fast mode*: `isFast` is set to `True`. This is the default mode for the notebooks, which means we train for fewer iterations or train / test on limited data. This ensures functional correctness of the notebook though the models produced are far from what a completed training would produce.\n",
"\n",
"- *Slow mode*: We recommend the user to set this flag to `False` once the user has gained familiarity with the notebook content and wants to gain insight from running the notebooks for a longer period with different parameters for training. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"isFast = True"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Reader\n",
"\n",
"To efficiently collect our data, randomize it for training, and pass it to the network, we use the CNTKTextFormat reader. We will create a small function that will be called when training (or testing) that defines the names of the streams in our data, and how they are referred to in the raw training data."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"# Helper function to load the model vocabulary file\n",
"def get_vocab(path):\n",
" # get the vocab for printing output sequences in plaintext\n",
" vocab = [w.strip() for w in open(path).readlines()]\n",
" i2w = { i:ch for i,ch in enumerate(vocab) }\n",
"\n",
" return (vocab, i2w)\n",
"\n",
"# Read vocabulary data and generate their corresponding indices\n",
"vocab, i2w = get_vocab(vocab_file)\n",
"\n",
"input_vocab_size = len(vocab)\n",
"label_vocab_size = len(vocab)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# Print vocab and the correspoding mapping to the phonemes\n",
"print(\"Vocabulary size is\", len(vocab))\n",
"print(\"First 15 letters are:\")\n",
"print(vocab[:15])\n",
"print()\n",
"print(\"Print dictionary with the vocabulary mapping:\")\n",
"print(i2w)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We will use the above to create a reader for our training data. Let's create it now:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"def create_reader(path, randomize, size=INFINITELY_REPEAT):\n",
" return MinibatchSource(CTFDeserializer(path, StreamDefs(\n",
" features = StreamDef(field='S0', shape=input_vocab_size, is_sparse=True),\n",
" labels = StreamDef(field='S1', shape=label_vocab_size, is_sparse=True)\n",
" )), randomize=randomize, epoch_size = size)\n",
"\n",
"# Train data reader\n",
"train_reader = create_reader(train_file, True)\n",
"\n",
"# Validation/Test data reader\n",
"valid_reader = create_reader(valid_file, False)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Now let's set our model hyperparameters..."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Our input vocabulary size is 69, and those ones represent the label as well. Additionally we have 1 hidden layer with 128 nodes."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"model_dir = \".\" # we downloaded our data to the local directory above # TODO check me\n",
"\n",
"# model dimensions\n",
"input_vocab_dim = input_vocab_size\n",
"label_vocab_dim = label_vocab_size\n",
"hidden_dim = 128\n",
"num_layers = 1"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Step 1: setup the input to the network\n",
"\n",
"### Dynamic axes in CNTK (Key concept)\n",
"\n",
"One of the important concepts in understanding CNTK is the idea of two types of axes:\n",
"- **static axes**, which are the traditional axes of a variable's shape, and\n",
"- **dynamic axes**, which have dimensions that are unknown until the variable is bound to real data at computation time.\n",
"\n",
"The dynamic axes are particularly important in the world of recurrent neural networks. Instead of having to decide a maximum sequence length ahead of time, padding your sequences to that size, and wasting computation, CNTK's dynamic axes allow for variable sequence lengths that are automatically packed in minibatches to be as efficient as possible.\n",
"\n",
"When setting up sequences, there are *two dynamic axes* that are important to consider. The first is the *batch axis*, which is the axis along which multiple sequences are batched. The second is the dynamic axis particular to that sequence. The latter is specific to a particular input because of variable sequence lengths in your data. For example, in sequence to sequence networks, we have two sequences: the **input sequence**, and the **output (or 'label') sequence**. One of the things that makes this type of network so powerful is that the length of the input sequence and the output sequence do not have to correspond to each other. Therefore, both the input sequence and the output sequence require their own unique dynamic axis.\n",
"\n",
"When defining the input to a network, we set up the required dynamic axes and the shape of the input variables. Below, we define the shape (vocabulary size) of the inputs, create their dynamic axes, and finally create input variables that represent input nodes in our network."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# Source and target inputs to the model\n",
"batch_axis = Axis.default_batch_axis()\n",
"input_seq_axis = Axis('inputAxis')\n",
"label_seq_axis = Axis('labelAxis')\n",
"\n",
"input_dynamic_axes = [batch_axis, input_seq_axis]\n",
"raw_input = input_variable(shape=(input_vocab_dim), dynamic_axes=input_dynamic_axes, name='raw_input')\n",
"\n",
"label_dynamic_axes = [batch_axis, label_seq_axis]\n",
"raw_labels = input_variable(shape=(label_vocab_dim), dynamic_axes=label_dynamic_axes, name='raw_labels')"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Questions\n",
"\n",
"1. Why do the shapes of the input variables correspond to the size of our dictionaries in sequence to sequence networks?"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Step 2: define the network\n",
"\n",
"As discussed before, the sequence-to-sequence network is, at its most basic, an RNN encoder followed by an RNN decoder, and a dense output layer. We could do this in a few lines with the layers library, but let's go through things in a little more detail without adding too much complexity. The first step is to perform some manipulations on the input data; let's look at the code below and then discuss what we're doing. "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# Instantiate the sequence to sequence translation model\n",
"input_sequence = raw_input\n",
"\n",
"# Drop the sentence start token from the label, for decoder training\n",
"label_sequence = sequence.slice(raw_labels,\n",
" 1, 0, name='label_sequence') # A B C --> A B C \n",
"label_sentence_start = sequence.first(raw_labels) # \n",
"\n",
"is_first_label = sequence.is_first(label_sequence) # 1 0 0 0 ...\n",
"label_sentence_start_scattered = sequence.scatter( # 0 0 0 ... (up to the length of label_sequence)\n",
" label_sentence_start, is_first_label)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We have two input variables, `raw_input` and `raw_labels`. Typically, the labels would not have to be part of the network definition because they would only be used in a criterion node when we compare the network's output with the ground truth. However, in sequence-to-sequence networks, the labels themselves form part of the input to the network during training as they are fed as the input into the decoder.\n",
"\n",
"To make use of these input variables, we will pass them through computation nodes. We first set `input_sequence` to `raw_input` as a convenience step. We then perform several modifications to `label_sequence` so that it will work with our network. For now you'll just have to trust that we will make good use of this stuff later.\n",
"\n",
"First, we slice the first element off of `label_sequence` so that it's missing the sentence-start token. This is because the decoder will always first be primed with that token, both during training and evaluation. When the ground truth isn't fed into the decoder, we will still feed in a sentence-start token, so we want to consistently view the input to the decoder as a sequence that starts with an actual value.\n",
"\n",
"Then, we get `label_sequence_start` by getting the `first` element from the sequence `raw_labels`. This will be used to compose a sequence that is the first input to the decoder regardless of whether we're training or decoding. Finally, the last two statements set up an actual sequence, with the correct dynamic axis, to be fed into the decoder. The function `sequence.scatter` takes the contents of `label_sentence_start` (which is ``) and turns it into a sequence with the first element containing the sequence start symbol and the rest of the elements containing 0's."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Let's create the LSTM recurrence"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"def LSTM_layer(input, output_dim, recurrence_hook_h=past_value, recurrence_hook_c=past_value):\n",
" # we first create placeholders for the hidden state and cell state which we don't have yet\n",
" dh = placeholder_variable(shape=(output_dim), dynamic_axes=input.dynamic_axes)\n",
" dc = placeholder_variable(shape=(output_dim), dynamic_axes=input.dynamic_axes)\n",
"\n",
" # we now create an LSTM_cell function and call it with the input and placeholders\n",
" LSTM_cell = LSTM(output_dim)\n",
" f_x_h_c = LSTM_cell(input, (dh, dc))\n",
" h_c = f_x_h_c.outputs\n",
"\n",
" # we setup the recurrence by specifying the type of recurrence (by default it's `past_value` -- the previous value)\n",
" h = recurrence_hook_h(h_c[0])\n",
" c = recurrence_hook_c(h_c[1])\n",
"\n",
" replacements = { dh: h.output, dc: c.output }\n",
" f_x_h_c.replace_placeholders(replacements)\n",
"\n",
" h = f_x_h_c.outputs[0]\n",
" c = f_x_h_c.outputs[1]\n",
"\n",
" # and finally we return the hidden state and cell state as functions (by using `combine`)\n",
" return combine([h]), combine([c])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Exercise 1: Create the encoder\n",
"\n",
"We will use the LSTM recurrence that we defined just above. Remember that its function signature is:\n",
"\n",
"`def LSTM_layer(input, output_dim, recurrence_hook_h=past_value, recurrence_hook_c=past_value):`\n",
"\n",
"and it returns a tuple `(hidden_state, hidden_cell)`. We will complete the following four exercises below. If possible, try them out before looking at the answers.\n",
"\n",
"1. Create the encoder (set the `output_dim` and `cell_dim` to `hidden_dim` which we defined earlier).\n",
"2. Set `num_layers` to something higher than 1 and create a stack of LSTMs to represent the encoder.\n",
"3. Get the output of the encoder and put it into the right form to be passed into the decoder [hard]\n",
"4. Reverse the order of the `input_sequence` (this has been shown to help especially in machine translation)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"# 1.\n",
"# Create the encoder (set the output_dim to hidden_dim which we defined earlier).\n",
"\n",
"(encoder_output_h, encoder_output_c) = LSTM_layer(input_sequence, hidden_dim)\n",
"\n",
"# 2.\n",
"# Set num_layers to something higher than 1 and create a stack of LSTMs to represent the encoder.\n",
"num_layers = 2\n",
"output_h = alias(input_sequence) # get a copy of the input_sequence\n",
"for i in range(0, num_layers):\n",
" (output_h, output_c) = LSTM_layer(output_h.output, hidden_dim)\n",
"\n",
"# 3.\n",
"# Get the output of the encoder and put it into the right form to be passed into the decoder [hard]\n",
"thought_vector_h = sequence.first(output_h)\n",
"thought_vector_c = sequence.first(output_c)\n",
"\n",
"thought_vector_broadcast_h = sequence.broadcast_as(thought_vector_h, label_sequence)\n",
"thought_vector_broadcast_c = sequence.broadcast_as(thought_vector_c, label_sequence)\n",
"\n",
"# 4.\n",
"# Reverse the order of the input_sequence (this has been shown to help especially in machine translation)\n",
"(encoder_output_h, encoder_output_c) = LSTM_layer(input_sequence, hidden_dim, future_value, future_value)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Exercise 2: Create the decoder\n",
"\n",
"In our basic version of the sequence-to-sequence network, the decoder generates an output sequence given the input sequence by setting the initial state of the decoder to the final hidden state of the encoder. The hidden state is represented by a tuple `(encoder_h, encoder_c)` where `h` represents the output hidden state and `c` represents the value of the LSTM cell.\n",
"\n",
"Besides setting the initial state of the decoder, we also need to give the decoder LSTM some input. The first element will always be the special sequence start tag ``. After that, there are two ways that we want to wire up the decoder's input: one during training, and the other during evaluation (i.e. generating sequences on the trained network).\n",
"\n",
"For training, the input to the decoder is the output sequence from the training data, also known as the label(s) for the input sequence. During evaluation, we will instead redirect the output from the network back into the decoder as its history. Let's first set up the input for training..."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"decoder_input = element_select(is_first_label, label_sentence_start_scattered, past_value(label_sequence))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Above, we use the function `element_select` which will return one of two options given the condition `is_first_label`. Remember that we're working with sequences so when the decoder LSTM is run its input will be unrolled along with the network. The above allows us to to have a dynamic input that will return a specific element given what time step we're currently processing.\n",
"\n",
"Therefore, the `decoder_input` will be `label_sentence_start_scattered` (which is simply ``) when we are at the first time step, and otherwise it will return the `past_value` (i.e. the previous element given what time step we're currently at) of `label_sequence`.\n",
"\n",
"Next, we need to setup our actual decoder. Before, for the encoder, we did the following:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"(output_h, output_c) = LSTM_layer(input_sequence, hidden_dim,\n",
" recurrence_hook_h=past_value, recurrence_hook_c=past_value)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To be able to set the first hidden state of the decoder to be equal to the final hidden state of the encoder, we can leverage the parameters `recurrence_hookH` and `recurrent_hookC`. The default `past_value` is a function that returns, for time `t`, the element in the sequence at time `t-1`. See if you can figure out how to set that up.\n",
"\n",
"1. Create the recurrence hooks for the decoder LSTM.\n",
" * Hint: you'll have to create a `lambda operand:` and you will make use of the `is_first_label` mask we used earlier and the `thought_vector_broadcast_h` and `thought_vector_broadcast_c` representations of the output of the encoder.\n",
"\n",
"2. With your recurrence hooks, create the decoder.\n",
" * Hint: again we'll use the `LSTMP_component_with_self_stabilization()` function and again use `hidden_dim` for the `output_dim` and `cell_dim`.\n",
"\n",
"3. Create a decoder with multiple layers. Note that you will have to use different recurrence hooks for the lower layers that feed back into the stack of layers."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"# 1.\n",
"# Create the recurrence hooks for the decoder LSTM.\n",
"\n",
"recurrence_hook_h = lambda operand: element_select(is_first_label, thought_vector_broadcast_h, past_value(operand))\n",
"recurrence_hook_c = lambda operand: element_select(is_first_label, thought_vector_broadcast_c, past_value(operand))\n",
"\n",
"# 2.\n",
"# With your recurrence hooks, create the decoder.\n",
"\n",
"(decoder_output_h, decoder_output_c) = LSTM_layer(decoder_input, hidden_dim, recurrence_hook_h, recurrence_hook_c)\n",
"\n",
"# 3.\n",
"# Create a decoder with multiple layers.\n",
"# Note that you will have to use different recurrence hooks for the lower layers\n",
"\n",
"num_layers = 3\n",
"decoder_output_h = alias(decoder_input)\n",
"for i in range(0, num_layers):\n",
" if (i > 0):\n",
" recurrence_hook_h = past_value\n",
" recurrence_hook_c = past_value\n",
" else:\n",
" recurrence_hook_h = lambda operand: element_select(\n",
" is_first_label, thought_vector_broadcast_h, past_value(operand))\n",
" recurrence_hook_c = lambda operand: element_select(\n",
" is_first_label, thought_vector_broadcast_c, past_value(operand))\n",
"\n",
" (decoder_output_h, decoder_output_c) = LSTM_layer(decoder_output_h.output, hidden_dim,\n",
" recurrence_hook_h, recurrence_hook_c)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Exercise 3: Fully connected layer (network output)\n",
"\n",
"Now we're almost at the end of defining the network. All we need to do is take the output of the decoder, and run it through a linear layer. Ultimately it will be put into a `softmax` to get a probability distribution over the possible output words. However, we will include that as part of our criterion nodes (below).\n",
"\n",
"1. Add the linear layer (a weight matrix, a bias parameter, a times, and a plus) to get the final output of the network"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"# 1.\n",
"# Add the linear layer\n",
"\n",
"W = parameter(shape=(decoder_output_h.shape[0], label_vocab_dim), init=glorot_uniform())\n",
"B = parameter(shape=(label_vocab_dim), init=0)\n",
"z = plus(B, times(decoder_output_h, W))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Putting the model together\n",
"\n",
"With the above we have defined some of the network and asked you to define parts of it as exercises. Here let's put the whole thing into a function called `create_model()`. Remember, all this does is create a skeleton of the network that defines how data will flow through it. No data is running through it yet."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"def create_model():\n",
"\n",
" # Source and target inputs to the model\n",
" batch_axis = Axis.default_batch_axis()\n",
" input_seq_axis = Axis('inputAxis')\n",
" label_seq_axis = Axis('labelAxis')\n",
"\n",
" input_dynamic_axes = [batch_axis, input_seq_axis]\n",
" raw_input = input_variable(\n",
" shape=(input_vocab_dim), dynamic_axes=input_dynamic_axes, name='raw_input')\n",
"\n",
" label_dynamic_axes = [batch_axis, label_seq_axis]\n",
" raw_labels = input_variable(\n",
" shape=(label_vocab_dim), dynamic_axes=label_dynamic_axes, name='raw_labels')\n",
"\n",
" # Instantiate the sequence to sequence translation model\n",
" input_sequence = raw_input\n",
"\n",
" # Drop the sentence start token from the label, for decoder training\n",
" label_sequence = sequence.slice(raw_labels, 1, 0,\n",
" name='label_sequence') # A B C --> A B C \n",
" label_sentence_start = sequence.first(raw_labels) # \n",
"\n",
" # Setup primer for decoder\n",
" is_first_label = sequence.is_first(label_sequence) # 1 0 0 0 ...\n",
" label_sentence_start_scattered = sequence.scatter(\n",
" label_sentence_start, is_first_label)\n",
"\n",
" # Encoder\n",
" stabilize = Stabilizer()\n",
" encoder_output_h = stabilize(input_sequence)\n",
" for i in range(0, num_layers):\n",
" (encoder_output_h, encoder_output_c) = LSTM_layer(\n",
" encoder_output_h.output, hidden_dim, future_value, future_value)\n",
"\n",
" # Prepare encoder output to be used in decoder\n",
" thought_vector_h = sequence.first(encoder_output_h)\n",
" thought_vector_c = sequence.first(encoder_output_c)\n",
"\n",
" thought_vector_broadcast_h = sequence.broadcast_as(\n",
" thought_vector_h, label_sequence)\n",
" thought_vector_broadcast_c = sequence.broadcast_as(\n",
" thought_vector_c, label_sequence)\n",
"\n",
" # Decoder\n",
" decoder_history_hook = alias(label_sequence, name='decoder_history_hook') # copy label_sequence\n",
"\n",
" decoder_input = element_select(is_first_label, label_sentence_start_scattered, past_value(\n",
" decoder_history_hook))\n",
"\n",
" decoder_output_h = stabilize(decoder_input)\n",
" for i in range(0, num_layers):\n",
" if (i > 0):\n",
" recurrence_hook_h = past_value\n",
" recurrence_hook_c = past_value\n",
" else:\n",
" recurrence_hook_h = lambda operand: element_select(\n",
" is_first_label, thought_vector_broadcast_h, past_value(operand))\n",
" recurrence_hook_c = lambda operand: element_select(\n",
" is_first_label, thought_vector_broadcast_c, past_value(operand))\n",
"\n",
" (decoder_output_h, decoder_output_c) = LSTM_layer(\n",
" decoder_output_h.output, hidden_dim, recurrence_hook_h, recurrence_hook_c)\n",
"\n",
" # Linear output layer\n",
" W = parameter(shape=(decoder_output_h.shape[0], label_vocab_dim), init=glorot_uniform())\n",
" B = parameter(shape=(label_vocab_dim), init=0)\n",
" z = plus(B, times(stabilize(decoder_output_h), W))\n",
"\n",
" return z"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Training\n",
"\n",
"Now that we've created the model, we are ready to train the network and learn its parameters. For sequence-to-sequence networks, the loss we use is cross-entropy. Note that we have to find the `label_sequences` node from the model because it was defined in our network and we want to compare the model's predictions specifically to the outputs of that node."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"model = create_model()\n",
"label_sequence = model.find_by_name('label_sequence')\n",
"\n",
"# Criterion nodes\n",
"ce = cross_entropy_with_softmax(model, label_sequence)\n",
"errs = classification_error(model, label_sequence)\n",
"\n",
"# let's show the required arguments for this model\n",
"print([x.name for x in model.arguments])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Next, we'll setup a bunch of parameters to drive our learning, we'll create the learner, and finally create our trainer:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# training parameters\n",
"lr_per_sample = learning_rate_schedule(0.007, UnitType.sample)\n",
"minibatch_size = 72\n",
"momentum_time_constant = momentum_as_time_constant_schedule(1100)\n",
"clipping_threshold_per_sample = 2.3\n",
"gradient_clipping_with_truncation = True\n",
"learner = momentum_sgd(model.parameters,\n",
" lr_per_sample, momentum_time_constant,\n",
" gradient_clipping_threshold_per_sample=clipping_threshold_per_sample,\n",
" gradient_clipping_with_truncation=gradient_clipping_with_truncation)\n",
"trainer = Trainer(model, ce, errs, learner)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"And now we bind the features and labels from our `train_reader` to the inputs that we setup in our network definition. First however, we'll define a convenience function to help find an argument name when pointing the reader's features to an argument of our model."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# helper function to find variables by name\n",
"def find_arg_by_name(name, expression):\n",
" vars = [i for i in expression.arguments if i.name == name]\n",
" assert len(vars) == 1\n",
" return vars[0]\n",
"\n",
"train_bind = {\n",
" find_arg_by_name('raw_input' , model) : train_reader.streams.features,\n",
" find_arg_by_name('raw_labels', model) : train_reader.streams.labels\n",
" }"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Finally, we define our training loop and start training the network!"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"training_progress_output_freq = 100\n",
"max_num_minibatch = 100 if isFast else 1000\n",
"\n",
"for i in range(max_num_minibatch):\n",
" # get next minibatch of training data\n",
" mb_train = train_reader.next_minibatch(minibatch_size, input_map=train_bind)\n",
" trainer.train_minibatch(mb_train)\n",
"\n",
" # collect epoch-wide stats\n",
" if i % training_progress_output_freq == 0:\n",
" print(\"Minibatch: {0}, Train Loss: {1:.3f}, Train Evaluation Criterion: {2:2.3f}\".format(i,\n",
" get_train_loss(trainer), get_train_eval_criterion(trainer)))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Model evaluation: greedy decoding\n",
"\n",
"Once we have a trained model, we of course then want to make use of it to generate output sequences! In this case, we will use greedy decoding. What this means is that we will run an input sequence through our trained network, and when we generate the output sequence, we will do so one element at a time by taking the `hardmax()` of the output of our network. This is obviously not optimal in general. Given the context, some word may always be the most probable at the first step, but another first word may be preferred given what is output later on. Decoding the optimal sequence is intractable in general. But we can do better doing a beam search where we keep around some small number of hypotheses at each step. However, greedy decoding can work surprisingly well for sequence-to-sequence networks because so much of the context is kept around in the RNN.\n",
"\n",
"To do greedy decoding, we need to hook in the previous output of our network as the input to the decoder. During training we passed the `label_sequences` (ground truth) in. You'll notice in our `create_model()` function above the following lines:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"decoder_history_hook = alias(label_sequence, name='decoder_history_hook') # copy label_sequence\n",
"decoder_input = element_select(is_first_label, label_sentence_start_scattered, past_value(decoder_history_hook))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This gives us a way to modify the `decoder_history_hook` after training to something else. We've already trained our network, but now we need a way to evaluate it without using a ground truth. We can do that like this:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": [
"model = create_model()\n",
"\n",
"# get some references to the new model\n",
"label_sequence = model.find_by_name('label_sequence')\n",
"decoder_history_hook = model.find_by_name('decoder_history_hook')\n",
"\n",
"# and now replace the output of decoder_history_hook with the hardmax output of the network\n",
"def clone_and_hook():\n",
" # network output for decoder history\n",
" net_output = hardmax(model)\n",
"\n",
" # make a clone of the graph where the ground truth is replaced by the network output\n",
" return model.clone(CloneMethod.share, {decoder_history_hook.output : net_output.output})\n",
"\n",
"# get a new model that uses the past network output as input to the decoder\n",
"new_model = clone_and_hook()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `new_model` now contains a version of the original network that shares parameters with it but that has a different input to the decoder. Namely, instead of feeding the ground truth labels into the decoder, it will feed in the history that the network has generated!\n",
"\n",
"Finally, let's see what it looks like if we train, and keep evaluating the network's output every `100` iterations by running a word's graphemes ('A B A D I') through our network. This way we can visualize the progress learning the best model... First we'll define a more complete `train()` action. It is largely the same as above but has some additional training parameters included; some additional smarts for printing out statistics as we go along; we now see progress over our data as epochs (one epoch is one complete pass over the training data); and we setup a reader for the single validation sequence we described above so that we can visually see our network's progress on that sequence as it learns."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"########################\n",
"# train action #\n",
"########################\n",
"\n",
"def train(train_reader, valid_reader, vocab, i2w, model, max_epochs):\n",
"\n",
" # do some hooks that we won't need in the future\n",
" label_sequence = model.find_by_name('label_sequence')\n",
" decoder_history_hook = model.find_by_name('decoder_history_hook')\n",
"\n",
" # Criterion nodes\n",
" ce = cross_entropy_with_softmax(model, label_sequence)\n",
" errs = classification_error(model, label_sequence)\n",
"\n",
" def clone_and_hook():\n",
" # network output for decoder history\n",
" net_output = hardmax(model)\n",
"\n",
" # make a clone of the graph where the ground truth is replaced by the network output\n",
" return model.clone(CloneMethod.share, {decoder_history_hook.output : net_output.output})\n",
"\n",
" # get a new model that uses the past network output as input to the decoder\n",
" new_model = clone_and_hook()\n",
"\n",
" # Instantiate the trainer object to drive the model training\n",
" lr_per_sample = learning_rate_schedule(0.007, UnitType.sample)\n",
" minibatch_size = 72\n",
" momentum_time_constant = momentum_as_time_constant_schedule(1100)\n",
" clipping_threshold_per_sample = 2.3\n",
" gradient_clipping_with_truncation = True\n",
" learner = momentum_sgd(model.parameters,\n",
" lr_per_sample, momentum_time_constant,\n",
" gradient_clipping_threshold_per_sample=clipping_threshold_per_sample,\n",
" gradient_clipping_with_truncation=gradient_clipping_with_truncation)\n",
" trainer = Trainer(model, ce, errs, learner)\n",
"\n",
" # Get minibatches of sequences to train with and perform model training\n",
" i = 0\n",
" mbs = 0\n",
"\n",
" # Set epoch size to a larger number of lower training error\n",
" epoch_size = 5000 if isFast else 908241\n",
"\n",
" training_progress_output_freq = 100\n",
"\n",
" # bind inputs to data from readers\n",
" train_bind = {\n",
" find_arg_by_name('raw_input' , model) : train_reader.streams.features,\n",
" find_arg_by_name('raw_labels', model) : train_reader.streams.labels\n",
" }\n",
" valid_bind = {\n",
" find_arg_by_name('raw_input' , new_model) : valid_reader.streams.features,\n",
" find_arg_by_name('raw_labels', new_model) : valid_reader.streams.labels\n",
" }\n",
"\n",
" for epoch in range(max_epochs):\n",
" loss_numer = 0\n",
" metric_numer = 0\n",
" denom = 0\n",
"\n",
" while i < (epoch+1) * epoch_size:\n",
" # get next minibatch of training data\n",
" mb_train = train_reader.next_minibatch(minibatch_size, input_map=train_bind)\n",
" trainer.train_minibatch(mb_train)\n",
"\n",
" # collect epoch-wide stats\n",
" samples = trainer.previous_minibatch_sample_count\n",
" loss_numer += trainer.previous_minibatch_loss_average * samples\n",
" metric_numer += trainer.previous_minibatch_evaluation_average * samples\n",
" denom += samples\n",
"\n",
" # every N MBs evaluate on a test sequence to visually show how we're doing; also print training stats\n",
" if mbs % training_progress_output_freq == 0:\n",
"\n",
" print(\"Minibatch: {0}, Train Loss: {1:2.3f}, Train Evaluation Criterion: {2:2.3f}\".format(mbs,\n",
" get_train_loss(trainer), get_train_eval_criterion(trainer)))\n",
"\n",
" mb_valid = valid_reader.next_minibatch(minibatch_size, input_map=valid_bind)\n",
" e = new_model.eval(mb_valid)\n",
" print_sequences(e, i2w)\n",
"\n",
" i += mb_train[find_arg_by_name('raw_labels', model)].num_samples\n",
" mbs += 1\n",
"\n",
" print(\"--- EPOCH %d DONE: loss = %f, errs = %f ---\" % (epoch, loss_numer/denom, 100.0*(metric_numer/denom)))\n",
" return 100.0*(metric_numer/denom)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Now that we have our three important functions defined -- `create_model()` and `train()`, let's make use of them:"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# Given a vocab and tensor, print the output\n",
"def print_sequences(sequences, i2w):\n",
" for s in sequences:\n",
" print([i2w[np.argmax(w)] for w in s], sep=\" \")\n",
"\n",
"# hook up data\n",
"train_reader = create_reader(train_file, True)\n",
"valid_reader = create_reader(valid_file, False)\n",
"vocab, i2w = get_vocab(vocab_file)\n",
"\n",
"# create model\n",
"model = create_model()\n",
"\n",
"# train\n",
"error = train(train_reader, valid_reader, vocab, i2w, model, max_epochs=1)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": false
},
"outputs": [],
"source": [
"# Print the training error\n",
"print(error)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Task\n",
"Note the error is very high. This is largely due to the minimum training we have done so far. Please change the `epoch_size` to be a much higher number and re-run the `train` function. This might take considerably longer time but you will see a marked reduction in the error."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Next steps\n",
"\n",
"An important extension to sequence-to-sequence models, especially when dealing with long sequences, is to use an attention mechanism. The idea behind attention is to allow the decoder, first, to look at any of the hidden state outputs from the encoder (instead of using only the final hidden state), and, second, to learn how much attention to pay to each of those hidden states given the context. This allows the outputted word at each time step `t` to depend not only on the final hidden state and the word that came before it, but instead on a weighted combination of *all* of the input hidden states!\n",
"\n",
"In the next version of this tutorial, we will talk about how to include attention in your sequence to sequence network."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"collapsed": true
},
"outputs": [],
"source": []
}
],
"metadata": {
"anaconda-cloud": {},
"kernelspec": {
"display_name": "Python [default]",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.4.5"
}
},
"nbformat": 4,
"nbformat_minor": 1
}
 \n",
"\n",
"In this tutorial, we are going to be talking about the fourth paradigm: many-to-many, also known as sequence-to-sequence networks. The input is a sequence with a dynamic length, and the output is also a sequence with some dynamic length. It is the logical extension of the many-to-one paradigm in that previously we were predicting some category (which could easily be one of `V` words where `V` is an entire vocabulary) and now we want to predict a whole sequence of those categories.\n",
"\n",
"The applications of sequence-to-sequence networks are nearly limitless. It is a natural fit for machine translation (e.g. English input sequences, French output sequences); automatic text summarization (e.g. full document input sequence, summary output sequence); word to pronunciation models (e.g. character [grapheme] input sequence, pronunciation [phoneme] output sequence); and even parse tree generation (e.g. regular text input, flat parse tree output).\n",
"\n",
"## Basic theory\n",
"\n",
"A sequence-to-sequence model consists of two main pieces: (1) an encoder; and (2) a decoder. Both the encoder and the decoder are recurrent neural network (RNN) layers that can be implemented using a vanilla RNN, an LSTM, or GRU cells (here we will use LSTM). In the basic sequence-to-sequence model, the encoder processes the input sequence into a fixed representation that is fed into the decoder as a context. The decoder then uses some mechanism (discussed below) to decode the processed information into an output sequence. The decoder is a language model that is augmented with some \"strong context\" by the encoder, and so each symbol that it generates is fed back into the decoder for additional context (like a traditional LM). For an English to German translation task, the most basic setup might look something like this:\n",
"\n",
"
\n",
"\n",
"In this tutorial, we are going to be talking about the fourth paradigm: many-to-many, also known as sequence-to-sequence networks. The input is a sequence with a dynamic length, and the output is also a sequence with some dynamic length. It is the logical extension of the many-to-one paradigm in that previously we were predicting some category (which could easily be one of `V` words where `V` is an entire vocabulary) and now we want to predict a whole sequence of those categories.\n",
"\n",
"The applications of sequence-to-sequence networks are nearly limitless. It is a natural fit for machine translation (e.g. English input sequences, French output sequences); automatic text summarization (e.g. full document input sequence, summary output sequence); word to pronunciation models (e.g. character [grapheme] input sequence, pronunciation [phoneme] output sequence); and even parse tree generation (e.g. regular text input, flat parse tree output).\n",
"\n",
"## Basic theory\n",
"\n",
"A sequence-to-sequence model consists of two main pieces: (1) an encoder; and (2) a decoder. Both the encoder and the decoder are recurrent neural network (RNN) layers that can be implemented using a vanilla RNN, an LSTM, or GRU cells (here we will use LSTM). In the basic sequence-to-sequence model, the encoder processes the input sequence into a fixed representation that is fed into the decoder as a context. The decoder then uses some mechanism (discussed below) to decode the processed information into an output sequence. The decoder is a language model that is augmented with some \"strong context\" by the encoder, and so each symbol that it generates is fed back into the decoder for additional context (like a traditional LM). For an English to German translation task, the most basic setup might look something like this:\n",
"\n",
" \n",
"\n",
"The basic sequence-to-sequence network passes the information from the encoder to the decoder by initializing the decoder RNN with the final hidden state of the encoder as its initial hidden state. The input is then a \"sequence start\" tag (`
\n",
"\n",
"The basic sequence-to-sequence network passes the information from the encoder to the decoder by initializing the decoder RNN with the final hidden state of the encoder as its initial hidden state. The input is then a \"sequence start\" tag (`