https://github.com/guifengwei/ChromHMM_mESC_mm10
Tip revision: 96506cbb5cd6be8e2bab3dc39300404f78cd8234 authored by Guifeng Wei on 11 April 2018, 11:01:34 UTC
Update README.md
Update README.md
Tip revision: 96506cb
README.md
# ChromHMM_mESC_mm10
Chromatin States Map in mouse embryonic stem cell
Author: Guifeng Wei
Date: Dec 2015
###### How I generated chromatin state maps in mouse embryonic stem cell (mm10):
First of all, the ChIP-seq data (H3K4me1, H3K4me3, H3K27me3, H3K27ac, H3K9me3, H3K9ac, H3K36me3, CTCF, Nanog, Oct4) in E14 cells generated by ENCODE project were downloaded.
Then following the ChromHMM manual, to identify genomic regions enriched in specific combinations of histone modifications as described previously but without extension of the reads.
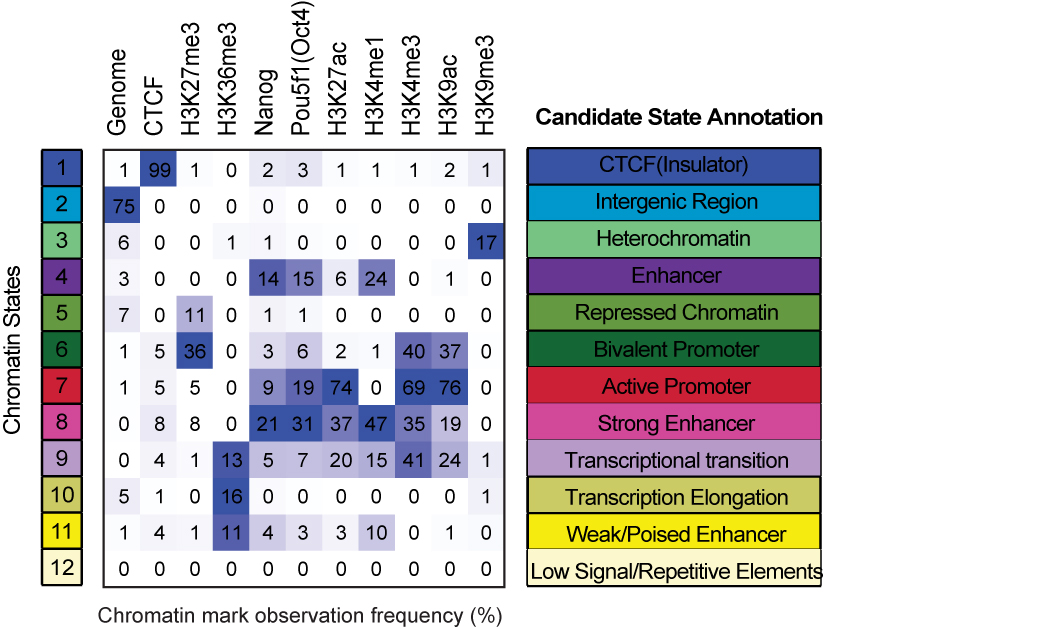
After testing n-state model from 9-15, finally I decided to use the 12-state model, which could interpret the chromatin frequency and validate the Nanog and Oct4 ChIP-seq properly.
The 12-state model contains the following states, mainly covering the promoter, enhancer, repressed Polycomb sites, transcription elongation region, CTCF bound sites and intergenic region.
These were the following commands that were used to generate maps of chromatin states in mouse
```javascript
java -mx4000M -jar ChromHMM.jar BinarizeBam mm10.chromsize.txt .. cellmarkfiletable mESC_E14_BinarizeBamDir/
```
```javascript
java -mx4000M -jar ChromHMM.jar LearnModel mESC_E14_BinarizeBamDir/ mESC_E14_ChromHMM_output/ 12 mm10
```
To interpret states, you can check this figure where I named each state with a specific color and indicated name:
##### Application:
There are many potential applications with the Chromatin States Map.
1. Annotate your ChIP-seq data to characterize which cis-elements in the mESC genome it prefer to occupy. You can use the command line with specified parameters.
```javascript
java -mx4000M -jar ChromHMM.jar OverlapEnrichment mESC_E14_12_segments.bed.gz
```
2. You can upload (mESC_E14_12_dense.annotated.bed) into your UCSC Genome Browser. Then you can browse your interest genomic regions or genes to see the around chromatin landscape. Here are some examples (the attached PDF file within this folder).
3. If you want to apply CRISPR to knockout certein DNA element, you can check whether the DNA element you would like to delete is functional in mESC.
##### Please Cite:
Pintacuda G.#, Wei G.#, Roustan C., Kirmitzas B.A., Solcan N., Cerase A., Castello A., Mohammed S., Moindrot B., Nesterova T.B. and Brockdorff N. (2017) hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Molecular Cell, 68, 955-969 e10.
##### Acknowledgments:
###### Jason Ernst http://www.biolchem.ucla.edu/labs/ernst/
###### ChromHMM http://compbio.mit.edu/ChromHMM/

