[](https://travis-ci.org/higlass/higlass)
[](https://docs.higlass.io/)
[](https://zenodo.org/badge/latestdoi/56026057)
[](https://twitter.com/higlass_io)
[](https://now-examples-slackin-zukqykuatj.now.sh/)
### Introduction
HiGlass is a web-based viewer for datasets too large to view at once.
It features synchronized navigation of multiple views as well as continuous zooming and panning
for navigation across genomic loci and resolutions. It supports visual comparison of
genomic (e.g., Hi-C, ChIP-seq, or bed annotations) and other data (e.g., geographic maps, gigapixel images, or abstract 1D and 2D sequential data) from different experimental conditions and can be used to efficiently
identify salient outcomes of experimental perturbations, generate new hypotheses, and share
the results with the community.
A live instance can be found at [http://higlass.io](http://higlass.io). A [Docker container](https://github.com/higlass/higlass-docker) is available for running an instance locally, although we recommend using the [higlass-manage](https://github.com/pkerpedjiev/higlass-manage) package to start, stop and configure local instances.
For documentation about how to use and install HiGlass, please visit [http://docs.higlass.io](http://docs.higlass.io).
### Citation
Kerpedjiev, P., Abdennur, N., Lekschas, F., McCallum, C., Dinkla, K., Strobelt, H., ... & Gehlenborg, N. *HiGlass: Web-based Visual Exploration and Analysis of Genome Interaction Maps.* Genome Biology (2018): 19:125. https://doi.org/10.1186/s13059-018-1486-1
### Example
<p align="center">
<img src="https://cloud.githubusercontent.com/assets/2143629/24535936/37ee60ee-15a5-11e7-89aa-434d93cda91d.gif" />
</p>
### Development
To run higlass from its source code simply run the following:
```
npm clean-install
npm run start
```
This starts a server in development mode at http://localhost:8080/.
Once started, a list of the examples can be found at [http://localhost:8080/examples.html](http://localhost:8080/examples.html).
Template viewconfs located at `/docs/examples/viewconfs` can viewed directly at urls such as [http://localhost:8080/apis/svg.html?/viewconfs/overlay-tracks.json](http://localhost:8080/apis/svg.html?/viewconfs/overlay-tracks.json).
### Tests
The tests for the React components and API functions are located in the `test` directory. To save time and only run relevant tests, open `karma.conf.js` and select the test files to run before running `test-watch`.
```
npm run test-watch
```
**Troubleshooting:**
- If the installation fails due to `sharp` > `node-gyp` try installing the node packages using `python2`:
```
npm ci --python=/usr/bin/python2 && rm -rf node_modules/node-sass && npm ci
```
### API
HiGlass provides an API for controlling the component from with JavaScript. Below is a [minimal working example](docs/examples/others/minimal-working-example.html) to get started and the complete documentation is availabe at [docs.higlass.io](http://docs.higlass.io/javascript_api.html).
```html
<!DOCTYPE html>
<head>
<meta charset="utf-8">
<title>Minimal Working Example · HiGlass</title>
<link rel="stylesheet" href="https://maxcdn.bootstrapcdn.com/bootstrap/3.3.7/css/bootstrap.min.css">
<link rel="stylesheet" href="https://unpkg.com/higlass@1.6.6/dist/hglib.css">
<style type="text/css">
html, body {
width: 100vw;
height: 100vh;
overflow: hidden;
}
</style>
<script crossorigin src="https://unpkg.com/react@16/umd/react.production.min.js"></script>
<script crossorigin src="https://unpkg.com/react-dom@16/umd/react-dom.production.min.js"></script>
<script crossorigin src="https://unpkg.com/pixi.js@5/dist/pixi.min.js"></script>
<script crossorigin src="https://unpkg.com/react-bootstrap@0.32.1/dist/react-bootstrap.min.js"></script>
<script crossorigin src="https://unpkg.com/higlass@1.6.6/dist/hglib.min.js"></script>
</head>
<body></body>
<script>
const hgApi = window.hglib.viewer(
document.body,
'https://higlass.io/api/v1/viewconfs/?d=default',
{ bounded: true },
);
</script>
</html>
```
### Related
[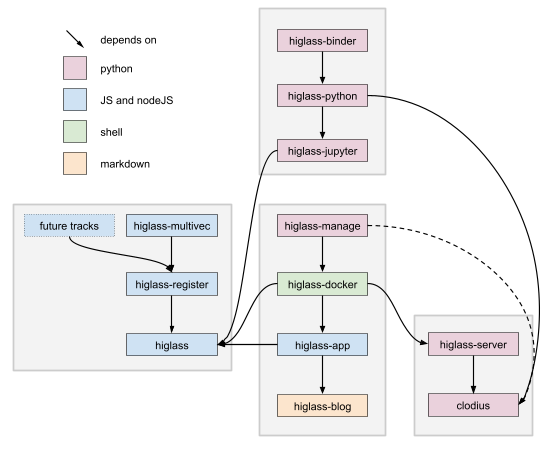](https://docs.google.com/drawings/d/1Xedi5ZRtbRdt2g20qpl_lWs4BMqc2DKZ2ZOoJvpHw9U/edit)
* [HiGlass Clodius](https://github.com/higlass/clodius) - Package that provides implementations for aggregation and tile generation for many common 1D and 2D data types
* [HiGlass Python](https://github.com/higlass/higlass-python) - Python bindings to the HiGlass for tile serving, view config generation, and Jupyter Notebook + Lab integration.
* [HiGlass Manage](https://github.com/higlass/higlass-manage) - Easy to use interface for deploying a local HiGlass instance
* [HiGlass Docker](https://github.com/higlass/higlass-docker) - Build an image containing all the components necessary to deploy HiGlass
* [HiGlass Server](https://github.com/higlass/higlass-server) - Server component for serving multi-resolution data
* [HiGlass App](https://github.com/higlass/higlass-app) - The code for the web application hosted at http://higlass.io
* [Cooler](https://github.com/mirnylab/cooler) - Package for efficient storage of and access to sparse 2D data
### License
HiGlass is provided under the MIT License.
