https://github.com/JuliaDiffEq/DiffEqFlux.jl
Tip revision: f4f9047659d68a7935f49fd26f86bfd1e637f30d authored by Chris Rackauckas on 20 October 2019, 18:27:43 UTC
Merge branch 'size'
Merge branch 'size'
Tip revision: f4f9047
README.md
# DiffEqFlux.jl
[](https://gitter.im/JuliaDiffEq/Lobby?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge)
[](https://travis-ci.org/JuliaDiffEq/DiffEqFlux.jl)
[](https://ci.appveyor.com/project/ChrisRackauckas/diffeqflux-jl)
[](https://gitlab.com/juliadiffeq/DiffEqFlux-jl/pipelines)
DiffEqFlux.jl fuses the world of differential equations with machine learning
by helping users put diffeq solvers into neural networks. This package utilizes
[DifferentialEquations.jl](http://docs.juliadiffeq.org/latest/) and
[Flux.jl](https://fluxml.ai/) as its building blocks to support research in
[Scientific Machine Learning](http://www.stochasticlifestyle.com/the-essential-tools-of-scientific-machine-learning-scientific-ml/)
and neural differential equations in traditional machine learning.
## Problem Domain
DiffEqFlux.jl is not just for neural ordinary differential equations. DiffEqFlux.jl is for neural differential equations.
As such, it is the first package to support and demonstrate:
- Stiff neural ordinary differential equations (neural ODEs)
- Neural stochastic differential equations (neural SDEs)
- Neural delay differential equations (neural DDEs)
- Neural partial differential equations (neural PDEs)
- Neural jump stochastic differential equations (neural jump diffusions)
with high order, adaptive, implicit, GPU-accelerated, Newton-Krylov, etc. methods. For examples, please refer to
[the release blog post](https://julialang.org/blog/2019/01/fluxdiffeq). Additional demonstrations, like neural
PDEs and neural jump SDEs, can be found [at this blog post](http://www.stochasticlifestyle.com/neural-jump-sdes-jump-diffusions-and-neural-pdes/) (among many others!).
Do not limit yourself to the current neuralization. With this package, you can explore various ways to integrate
the two methodologies:
- Neural networks can be defined where the “activations” are nonlinear functions described by differential equations.
- Neural networks can be defined where some layers are ODE solves
- ODEs can be defined where some terms are neural networks
- Cost functions on ODEs can define neural networks
## Citation
If you use DiffEqFlux.jl or are influenced by its ideas for expanding beyond neural ODEs, please cite:
```
@article{DBLP:journals/corr/abs-1902-02376,
author = {Christopher Rackauckas and
Mike Innes and
Yingbo Ma and
Jesse Bettencourt and
Lyndon White and
Vaibhav Dixit},
title = {DiffEqFlux.jl - {A} Julia Library for Neural Differential Equations},
journal = {CoRR},
volume = {abs/1902.02376},
year = {2019},
url = {http://arxiv.org/abs/1902.02376},
archivePrefix = {arXiv},
eprint = {1902.02376},
timestamp = {Tue, 21 May 2019 18:03:36 +0200},
biburl = {https://dblp.org/rec/bib/journals/corr/abs-1902-02376},
bibsource = {dblp computer science bibliography, https://dblp.org}
}
```
## Example Usage
For an overview of what this package is for, [see this blog post](https://julialang.org/blog/2019/01/fluxdiffeq).
### Optimizing parameters of an ODE
First let's create a Lotka-Volterra ODE using DifferentialEquations.jl. For
more details, [see the DifferentialEquations.jl documentation](http://docs.juliadiffeq.org/latest/)
```julia
using DifferentialEquations
function lotka_volterra(du,u,p,t)
x, y = u
α, β, δ, γ = p
du[1] = dx = α*x - β*x*y
du[2] = dy = -δ*y + γ*x*y
end
u0 = [1.0,1.0]
tspan = (0.0,10.0)
p = [1.5,1.0,3.0,1.0]
prob = ODEProblem(lotka_volterra,u0,tspan,p)
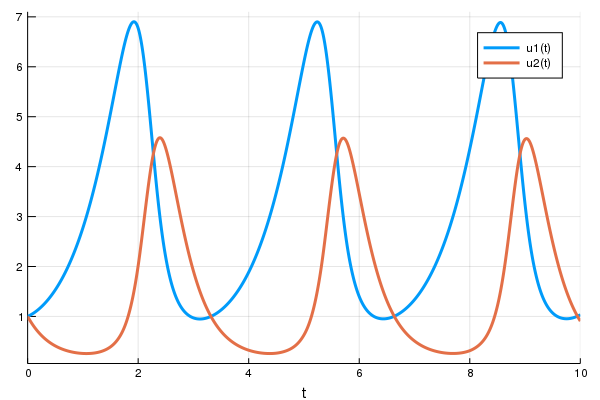
sol = solve(prob,Tsit5())
using Plots
plot(sol)
```
Next we define a single layer neural network that uses the `diffeq_adjoint` layer
function that takes the parameters and returns the solution of the `x(t)`
variable. Note that the `diffeq_adjoint` is usually preferred for ODEs, but does not
extend to other differential equation types (see the performance discussion section for
details). Instead of being a function of the parameters, we will wrap our
parameters in `param` to be tracked by Flux:
```julia
using Flux, DiffEqFlux
p = param([2.2, 1.0, 2.0, 0.4]) # Initial Parameter Vector
params = Flux.Params([p])
function predict_adjoint() # Our 1-layer neural network
diffeq_adjoint(p,prob,Tsit5(),saveat=0.0:0.1:10.0)
end
```
Next we choose a loss function. Our goal will be to find parameter that make
the Lotka-Volterra solution constant `x(t)=1`, so we defined our loss as the
squared distance from 1:
```julia
loss_adjoint() = sum(abs2,x-1 for x in predict_adjoint())
```
Lastly, we train the neural network using Flux to arrive at parameters which
optimize for our goal:
```julia
data = Iterators.repeated((), 100)
opt = ADAM(0.1)
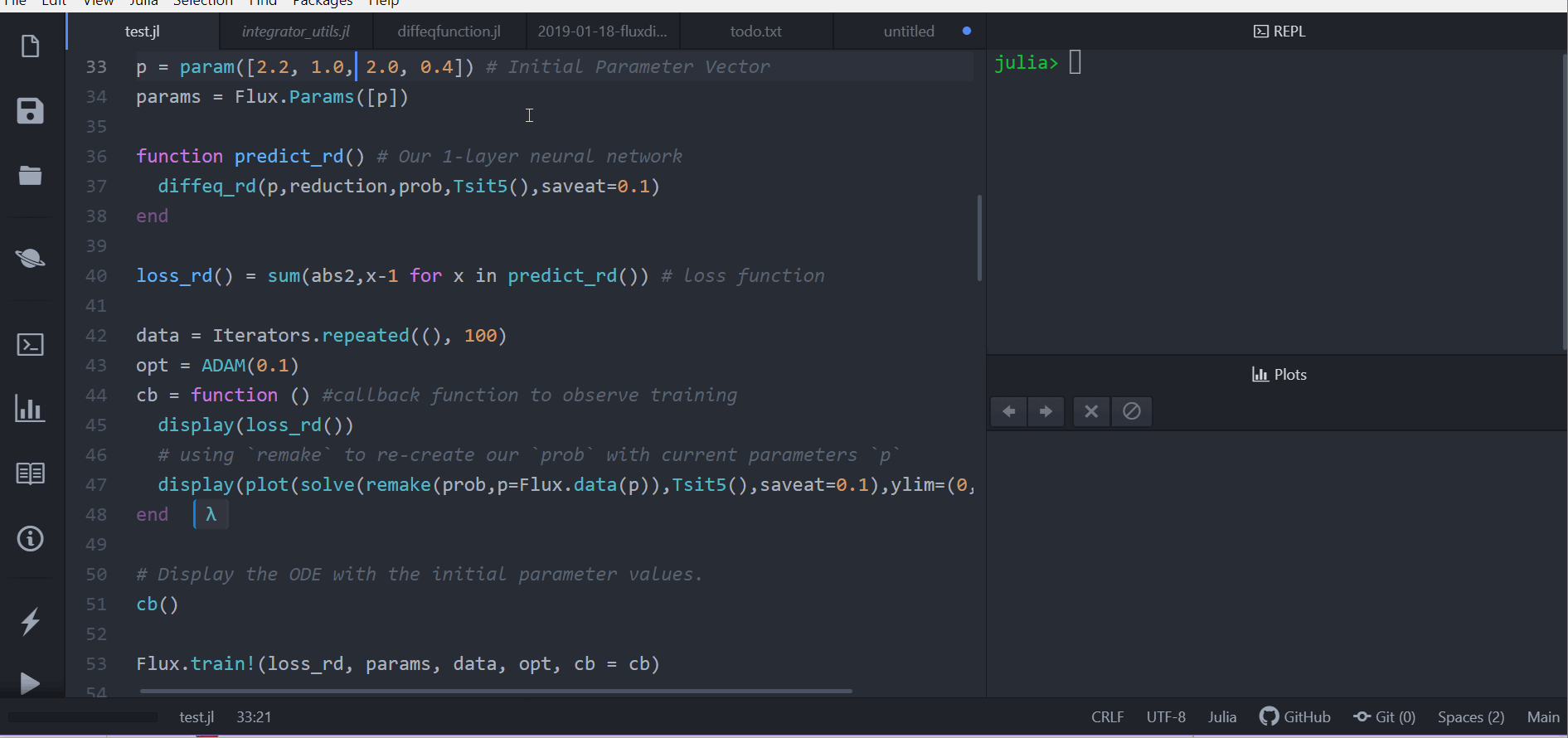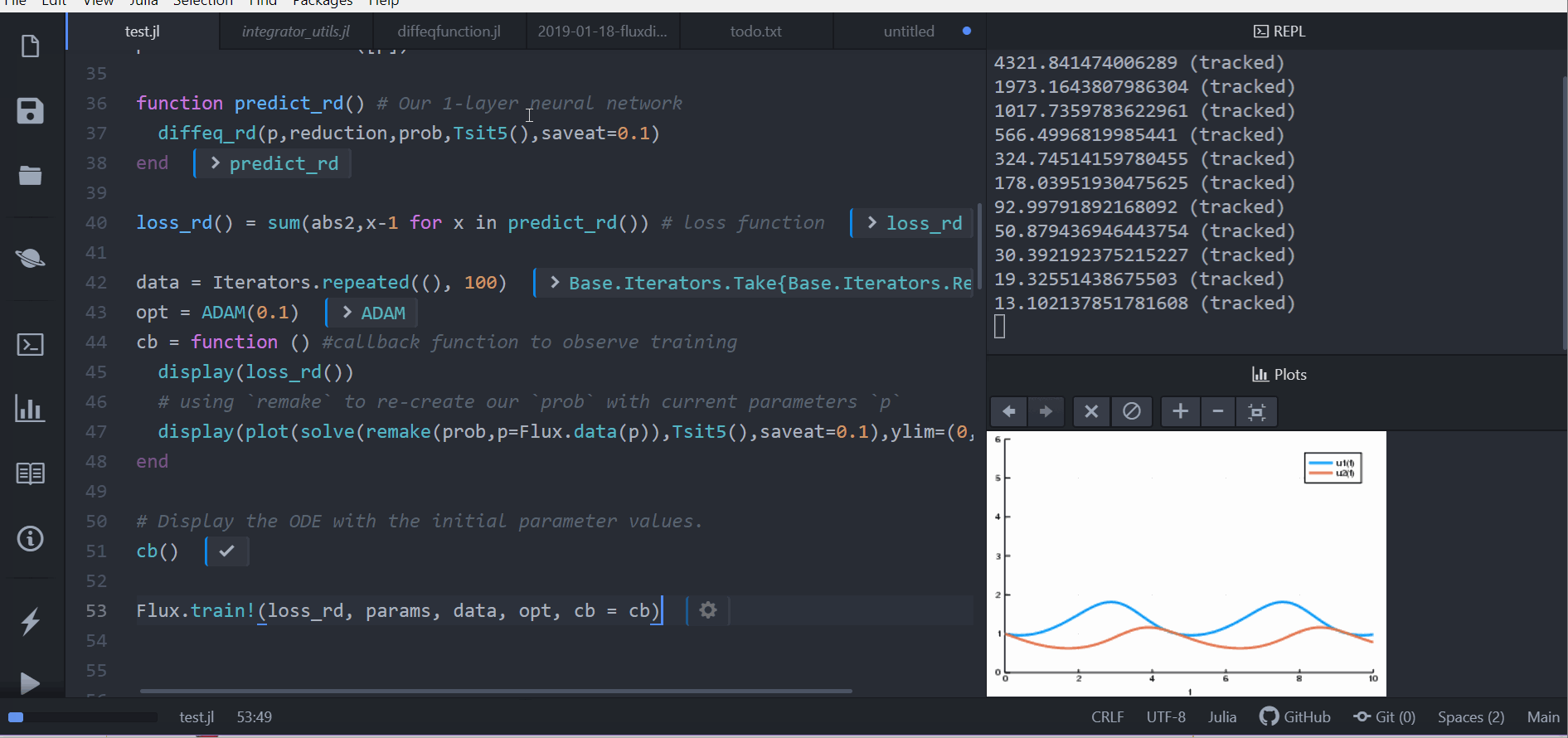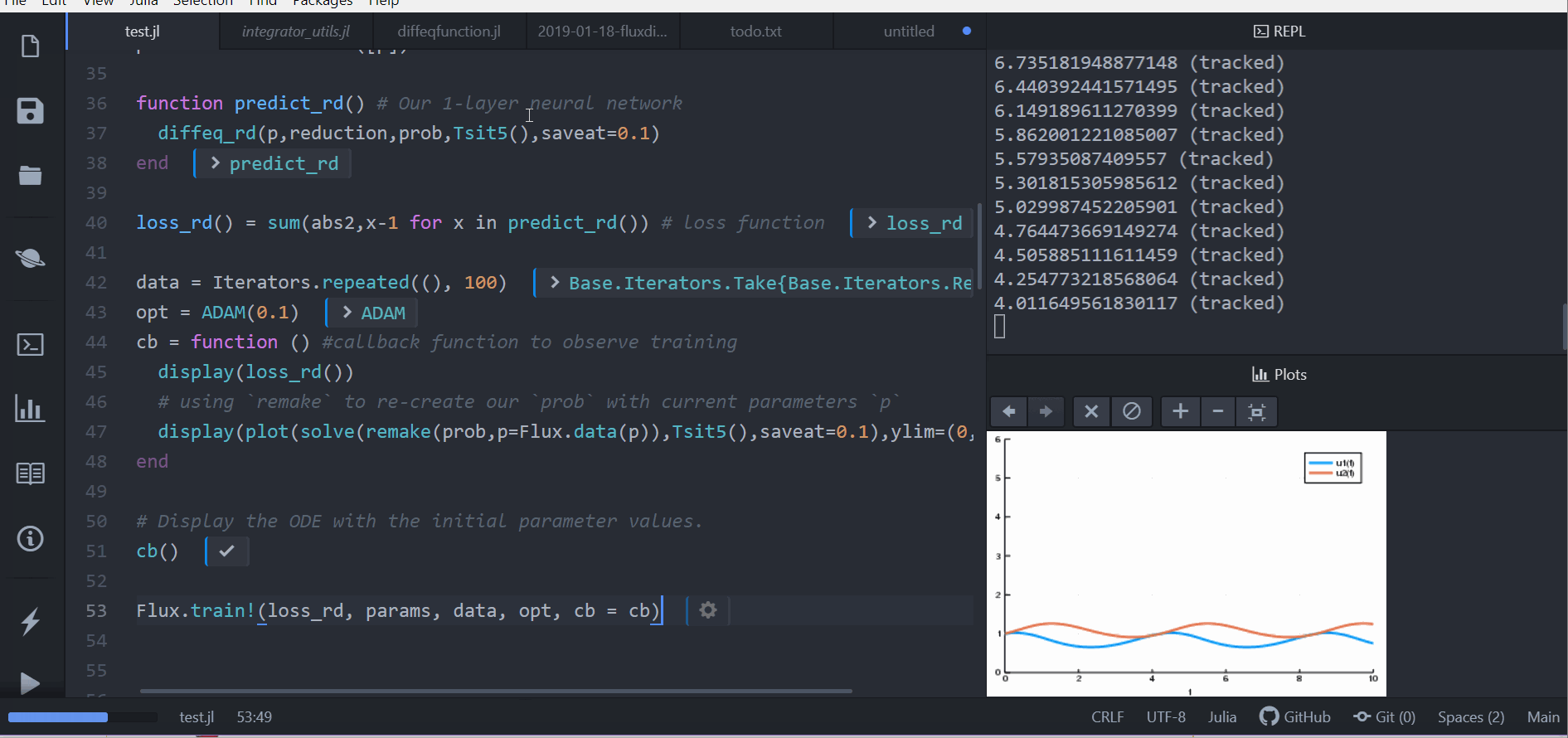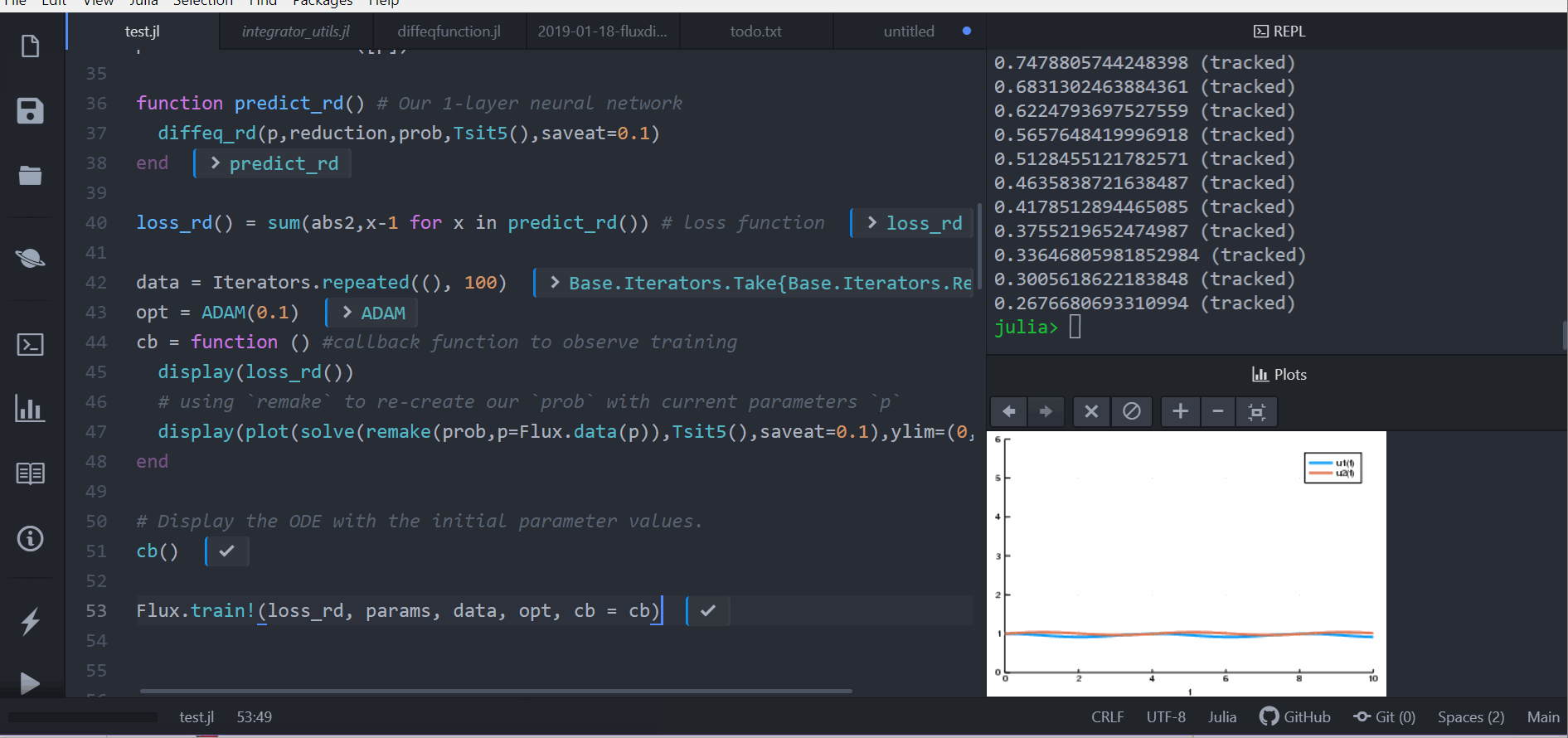
cb = function () #callback function to observe training
display(loss_adjoint())
# using `remake` to re-create our `prob` with current parameters `p`
display(plot(solve(remake(prob,p=Flux.data(p)),Tsit5(),saveat=0.0:0.1:10.0),ylim=(0,6)))
end
# Display the ODE with the initial parameter values.
cb()
Flux.train!(loss_adjoint, params, data, opt, cb = cb)
```
Note that by using anonymous functions, this `diffeq_adjoint` can be used as a
layer in a neural network `Chain`, for example like
```julia
m = Chain(
Conv((2,2), 1=>16, relu),
x -> maxpool(x, (2,2)),
Conv((2,2), 16=>8, relu),
x -> maxpool(x, (2,2)),
x -> reshape(x, :, size(x, 4)),
# takes in the ODE parameters from the previous layer
p -> diffeq_adjoint(p,prob,Tsit5(),saveat=0.1),
Dense(288, 10), softmax) |> gpu
```
or
```julia
m = Chain(
Dense(28^2, 32, relu),
# takes in the initial condition from the previous layer
x -> diffeq_rd(p,prob,Tsit5(),saveat=0.1,u0=x)),
Dense(32, 10),
softmax)
```
Similarly, `diffeq_adjoint`, a O(1) memory adjoint implementation, can be
replaced with `diffeq_rd` for reverse-mode automatic differentiation or
`diffeq_fd` for forward-mode automatic differentiation. `diffeq_fd` will
be fastest with small numbers of parameters, while `diffeq_adjoint` will
be the fastest when there are large numbers of parameters (like with a
neural ODE). See the layer API documentation for details.
### Using Other Differential Equations
Other differential equation problem types from DifferentialEquations.jl are
supported. For example, we can build a layer with a delay differential equation
like:
```julia
function delay_lotka_volterra(du,u,h,p,t)
x, y = u
α, β, δ, γ = p
du[1] = dx = (α - β*y)*h(p,t-0.1)[1]
du[2] = dy = (δ*x - γ)*y
end
h(p,t) = ones(eltype(p),2)
prob = DDEProblem(delay_lotka_volterra,[1.0,1.0],h,(0.0,10.0),constant_lags=[0.1])
p = param([2.2, 1.0, 2.0, 0.4])
params = Flux.Params([p])
function predict_rd_dde()
Array(diffeq_rd(p,prob,MethodOfSteps(Tsit5()),saveat=0.1))
end
loss_rd_dde() = sum(abs2,x-1 for x in predict_rd_dde())
loss_rd_dde()
```
Notice that we used mutating reverse-mode to handle a small delay differential
equation, a strategy that can be good for small equations (see the performance
discussion for more details on other forms).
Or we can use a stochastic differential equation. Here we demonstrate
`diffeq_fd` for forward-mode automatic differentiation of a small differential
equation:
```julia
function lotka_volterra_noise(du,u,p,t)
du[1] = 0.1u[1]
du[2] = 0.1u[2]
end
prob = SDEProblem(lotka_volterra,lotka_volterra_noise,[1.0,1.0],(0.0,10.0))
p = param([2.2, 1.0, 2.0, 0.4])
params = Flux.Params([p])
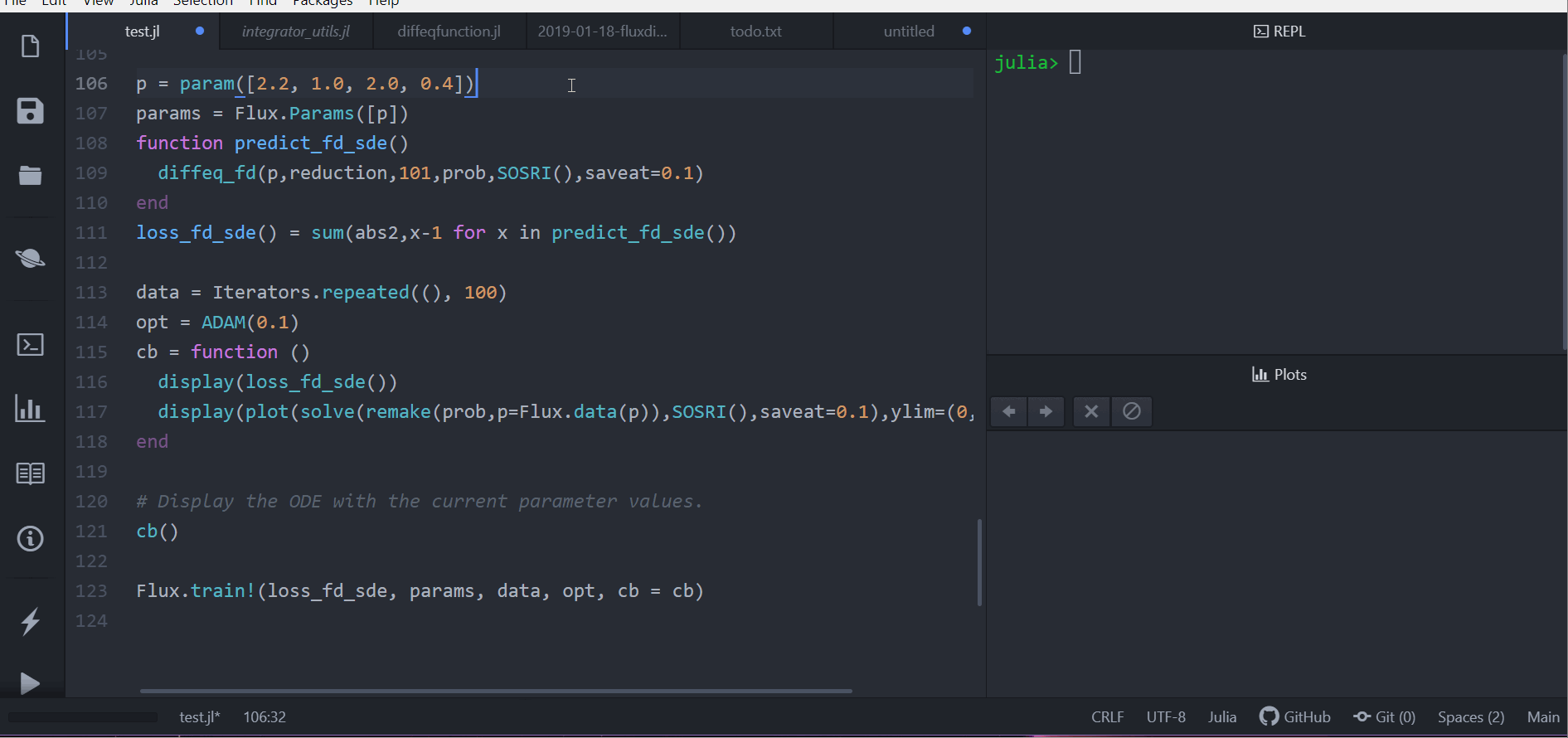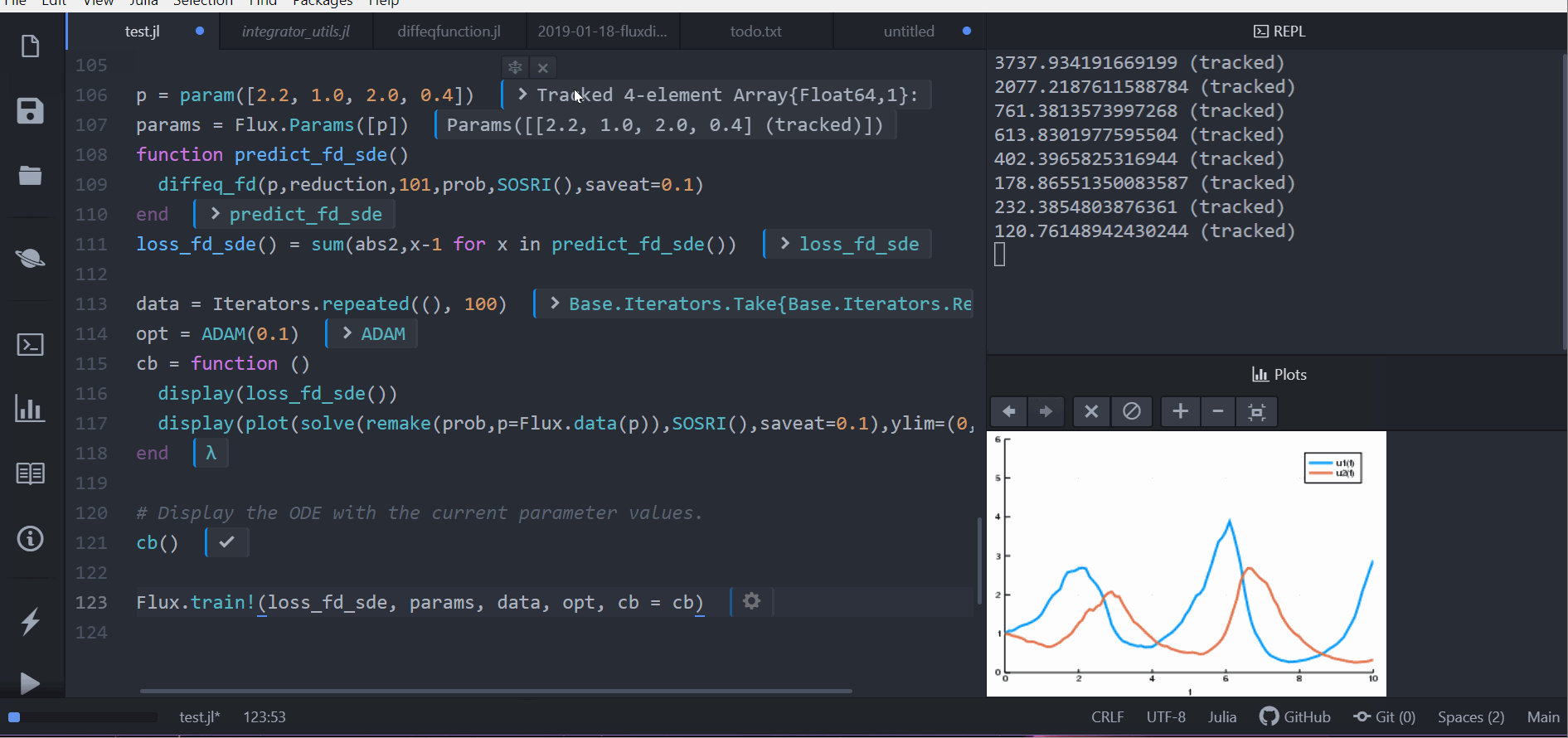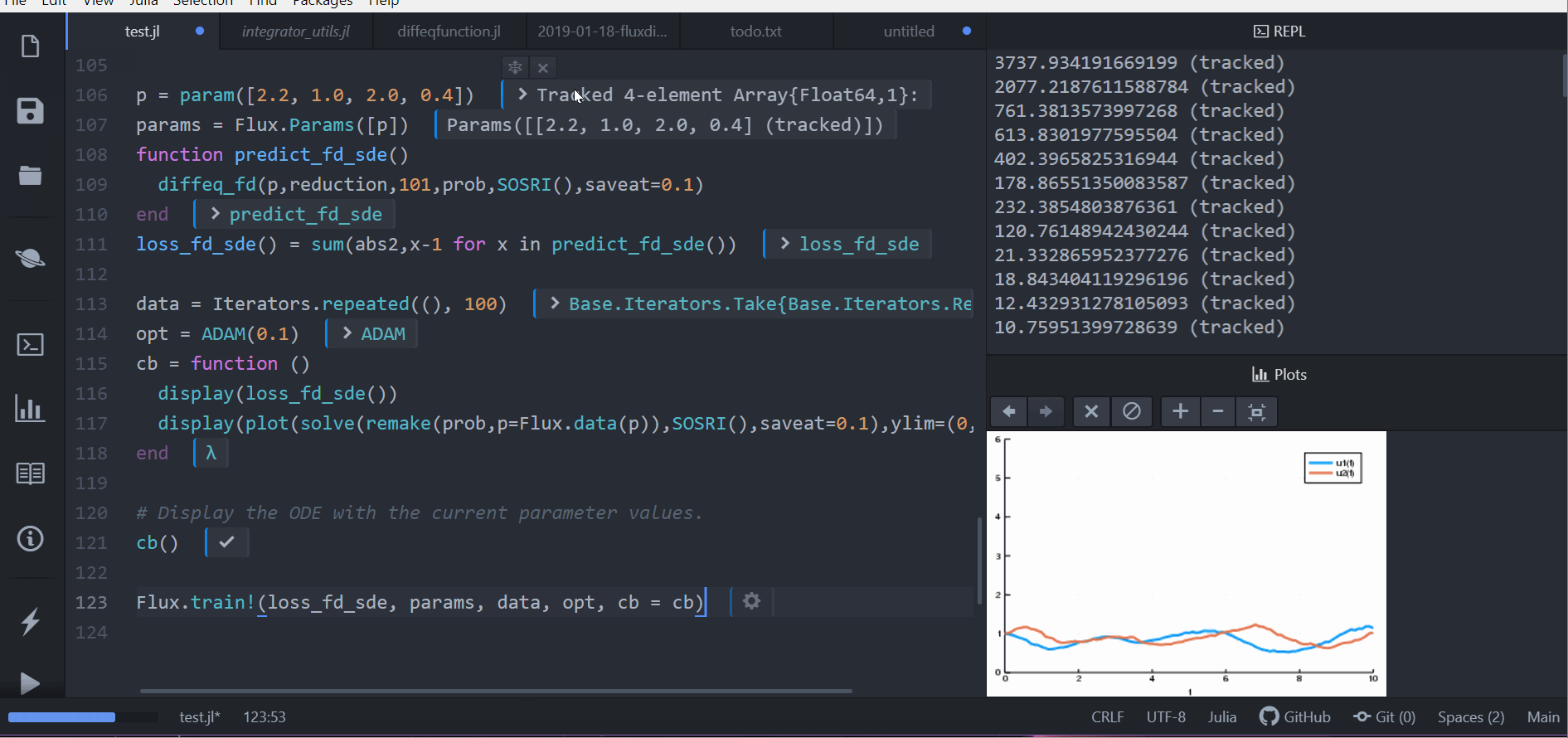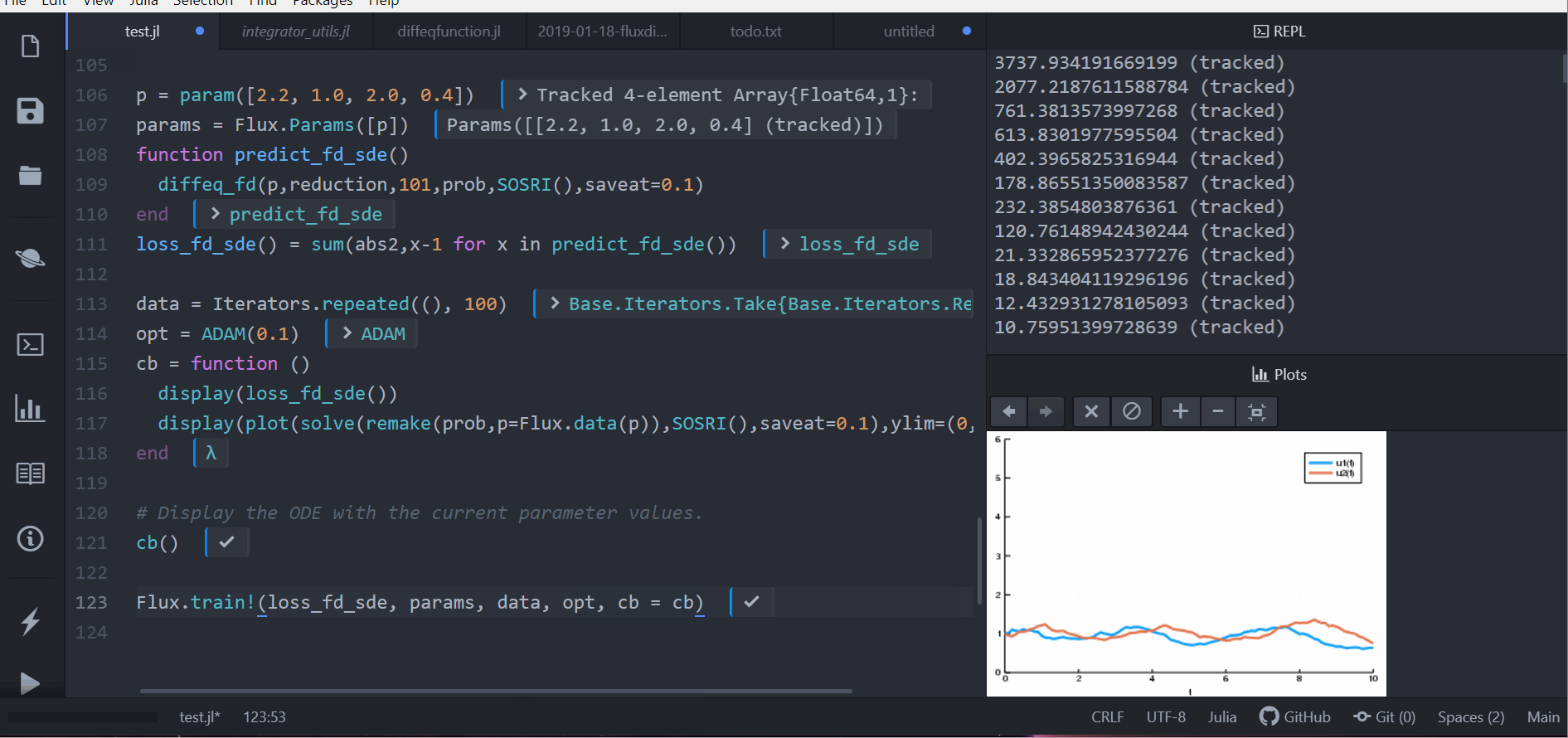
function predict_fd_sde()
diffeq_fd(p,Array,202,prob,SOSRI(),saveat=0.1)
end
loss_fd_sde() = sum(abs2,x-1 for x in predict_fd_sde())
loss_fd_sde()
data = Iterators.repeated((), 100)
opt = ADAM(0.1)
cb = function ()
display(loss_fd_sde())
display(plot(solve(remake(prob,p=Flux.data(p)),SOSRI(),saveat=0.1),ylim=(0,6)))
end
# Display the ODE with the current parameter values.
cb()
Flux.train!(loss_fd_sde, params, data, opt, cb = cb)
```
### Neural Ordinary Differential Equations
We can use DiffEqFlux.jl to define, solve, and train neural ordinary differential
equations. A neural ODE is an ODE where a neural network defines its derivative
function. Thus for example, with the multilayer perceptron neural network
`Chain(Dense(2,50,tanh),Dense(50,2))`, the best way to define a neural ODE by hand
would be to use non-mutating adjoints, which looks like:
```julia
p = DiffEqFlux.destructure(model)
dudt_(u::TrackedArray,p,t) = DiffEqFlux.restructure(model,p)(u)
dudt_(u::AbstractArray,p,t) = Flux.data(DiffEqFlux.restructure(model,p)(u))
prob = ODEProblem(dudt_,x,tspan,p)
my_neural_ode_prob = diffeq_adjoint(p,prob,args...;u0=x,kwargs...)
```
(`DiffEqFlux.restructure` and `DiffEqFlux.destructure` are helper functions
which transform the neural network to use parameters `p`)
A convenience function which handles all of the details is `neural_ode`. To
use `neural_ode`, you give it the initial condition, the internal neural
network model to use, the timespan to solve on, and any ODE solver arguments.
For example, this neural ODE would be defined as:
```julia
tspan = (0.0f0,25.0f0)
x -> neural_ode(model,x,tspan,Tsit5(),saveat=0.1)
```
where here we made it a layer that takes in the initial condition and spits
out an array for the time series saved at every 0.1 time steps.
### Training a Neural Ordinary Differential Equation
Let's get a time series array from the Lotka-Volterra equation as data:
```julia
u0 = Float32[2.; 0.]
datasize = 30
tspan = (0.0f0,1.5f0)
function trueODEfunc(du,u,p,t)
true_A = [-0.1 2.0; -2.0 -0.1]
du .= ((u.^3)'true_A)'
end
t = range(tspan[1],tspan[2],length=datasize)
prob = ODEProblem(trueODEfunc,u0,tspan)
ode_data = Array(solve(prob,Tsit5(),saveat=t))
```
Now let's define a neural network with a `neural_ode` layer. First we define
the layer:
```julia
dudt = Chain(x -> x.^3,
Dense(2,50,tanh),
Dense(50,2))
n_ode(x) = neural_ode(dudt,x,tspan,Tsit5(),saveat=t,reltol=1e-7,abstol=1e-9)
```
Here we used the `x -> x.^3` assumption in the model. By incorporating structure
into our equations, we can reduce the required size and training time for the
neural network, but a good guess needs to be known!
From here we build a loss function around it. We will use the L2 loss of the network's
output against the time series data:
```julia
function predict_n_ode()
n_ode(u0)
end
loss_n_ode() = sum(abs2,ode_data .- predict_n_ode())
```
and then train the neural network to learn the ODE:
```julia
data = Iterators.repeated((), 1000)
opt = ADAM(0.1)
cb = function () #callback function to observe training
display(loss_n_ode())
# plot current prediction against data
cur_pred = Flux.data(predict_n_ode())
pl = scatter(t,ode_data[1,:],label="data")
scatter!(pl,t,cur_pred[1,:],label="prediction")
display(plot(pl))
end
# Display the ODE with the initial parameter values.
cb()
ps = Flux.params(dudt)
Flux.train!(loss_n_ode, ps, data, opt, cb = cb)
```
## Use with GPUs
Note that the differential equation solvers will run on the GPU if the initial
condition is a GPU array. Thus for example, we can define a neural ODE by hand
that runs on the GPU:
```julia
u0 = Float32[2.; 0.] |> gpu
dudt = Chain(Dense(2,50,tanh),Dense(50,2)) |> gpu
p = DiffEqFlux.destructure(model)
dudt_(u::TrackedArray,p,t) = DiffEqFlux.restructure(model,p)(u)
dudt_(u::AbstractArray,p,t) = Flux.data(DiffEqFlux.restructure(model,p)(u))
prob = ODEProblem(ODEfunc, u0,tspan)
# Runs on a GPU
sol = solve(prob,Tsit5(),saveat=0.1)
```
and the `diffeq` layer functions can be used similarly. Or we can directly use
the neural ODE layer function, like:
```julia
x -> neural_ode(gpu(dudt),gpu(x),tspan,Tsit5(),saveat=0.1)
```
## Mixed Neural DEs
You can also mix a known differential equation and a neural differential equation, so that
the parameters and the neural network are estimated simultaniously. Here's an example of
doing this with both reverse-mode autodifferentiation and with adjoints:
```julia
using DiffEqFlux, Flux, OrdinaryDiffEq
## --- Reverse-mode AD ---
tspan = (0.0f0,25.0f0)
u0 = Tracker.param(Float32[0.8; 0.8])
ann = Chain(Dense(2,10,tanh), Dense(10,1))
p = param(Float32[-2.0,1.1])
function dudt_(u::TrackedArray,p,t)
x, y = u
Flux.Tracker.collect(
[ann(u)[1],
p[1]*y + p[2]*x*y])
end
function dudt_(u::AbstractArray,p,t)
x, y = u
[Flux.data(ann(u)[1]),
p[1]*y + p[2]*x*y]
end
prob = ODEProblem(dudt_,u0,tspan,p)
diffeq_rd(p,prob,Tsit5())
function predict_rd()
Flux.Tracker.collect(diffeq_rd(p,prob,Tsit5(),u0=u0))
end
loss_rd() = sum(abs2,x-1 for x in predict_rd())
loss_rd()
data = Iterators.repeated((), 10)
opt = ADAM(0.1)
cb = function ()
display(loss_rd())
#display(plot(solve(remake(prob,u0=Flux.data(u0),p=Flux.data(p)),Tsit5(),saveat=0.1),ylim=(0,6)))
end
# Display the ODE with the current parameter values.
cb()
Flux.train!(loss_rd, params(ann,p,u0), data, opt, cb = cb)
## --- Partial Neural Adjoint ---
u0 = param(Float32[0.8; 0.8])
tspan = (0.0f0,25.0f0)
ann = Chain(Dense(2,10,tanh), Dense(10,1))
p1 = Flux.data(DiffEqFlux.destructure(ann))
p2 = Float32[-2.0,1.1]
p3 = param([p1;p2])
ps = Flux.params(p3,u0)
function dudt_(du,u,p,t)
x, y = u
du[1] = DiffEqFlux.restructure(ann,p[1:41])(u)[1]
du[2] = p[end-1]*y + p[end]*x
end
prob = ODEProblem(dudt_,u0,tspan,p3)
diffeq_adjoint(p3,prob,Tsit5(),u0=u0,abstol=1e-8,reltol=1e-6)
function predict_adjoint()
diffeq_adjoint(p3,prob,Tsit5(),u0=u0,saveat=0.0:0.1:25.0)
end
loss_adjoint() = sum(abs2,x-1 for x in predict_adjoint())
loss_adjoint()
data = Iterators.repeated((), 10)
opt = ADAM(0.1)
cb = function ()
display(loss_adjoint())
#display(plot(solve(remake(prob,p=Flux.data(p3),u0=Flux.data(u0)),Tsit5(),saveat=0.1),ylim=(0,6)))
end
# Display the ODE with the current parameter values.
cb()
Flux.train!(loss_adjoint, ps, data, opt, cb = cb)
```
## Neural Differential Equations for Non-ODEs: Neural SDEs, Neural DDEs, etc.
With neural stochastic differential equations, there is once again a helper form `neural_dmsde` which can
be used for the multiplicative noise case (consult the layers API documentation, or
[this full example using the layer function](https://github.com/MikeInnes/zygote-paper/blob/master/neural_sde/neural_sde.jl)).
However, since there are far too many possible combinations for the API to support, in many cases you will want to
performantly define neural differential equations for non-ODE systems from scratch. For these systems, it is generally
best to use `diffeq_rd` with non-mutating (out-of-place) forms. For example, the following defines a neural SDE with
neural networks for both the drift and diffusion terms:
```julia
dudt_(u,p,t) = model(u)
g(u,p,t) = model2(u)
prob = SDEProblem(dudt_,g,param(x),tspan,nothing)
```
where `model` and `model2` are different neural networks. The same can apply to a neural delay differential equation.
Its out-of-place formulation is `f(u,h,p,t)`. Thus for example, if we want to define a neural delay differential equation
which uses the history value at `p.tau` in the past, we can define:
```julia
dudt_(u,h,p,t) = model([u;h(t-p.tau)])
prob = DDEProblem(dudt_,u0,h,tspan,nothing)
```
### Neural SDE Example
First let's build training data from the same example as the neural ODE:
```julia
using Flux, DiffEqFlux, StochasticDiffEq, Plots, DiffEqBase.EnsembleAnalysis
u0 = Float32[2.; 0.]
datasize = 30
tspan = (0.0f0,1.0f0)
function trueODEfunc(du,u,p,t)
true_A = [-0.1 2.0; -2.0 -0.1]
du .= ((u.^3)'true_A)'
end
t = range(tspan[1],tspan[2],length=datasize)
mp = Float32[0.2,0.2]
function true_noise_func(du,u,p,t)
du .= mp.*u
end
prob = SDEProblem(trueODEfunc,true_noise_func,u0,tspan)
```
For our dataset we will use DifferentialEquations.jl's [parallel ensemble interface](http://docs.juliadiffeq.org/latest/features/ensemble.html)
to generate data from the average of 100 runs of the SDE:
```julia
# Take a typical sample from the mean
ensemble_prob = EnsembleProblem(prob)
ensemble_sol = solve(ensemble_prob,SOSRI(),trajectories = 100)
ensemble_sum = EnsembleSummary(ensemble_sol)
sde_data = Array(timeseries_point_mean(ensemble_sol,t))
```
Now we build a neural SDE. For simplicity we will use the `neural_dmsde`
multiplicative noise neural SDE layer function:
```julia
dudt = Chain(x -> x.^3,
Dense(2,50,tanh),
Dense(50,2))
ps = Flux.params(dudt)
n_sde = x->neural_dmsde(dudt,x,mp,tspan,SOSRI(),saveat=t,reltol=1e-1,abstol=1e-1)
```
Let's see what that looks like:
```julia
pred = n_sde(u0) # Get the prediction using the correct initial condition
dudt_(u,p,t) = Flux.data(dudt(u))
g(u,p,t) = mp.*u
nprob = SDEProblem(dudt_,g,u0,(0.0f0,1.2f0),nothing)
ensemble_nprob = EnsembleProblem(nprob)
ensemble_nsol = solve(ensemble_nprob,SOSRI(),trajectories = 100)
ensemble_nsum = EnsembleSummary(ensemble_nsol)
p1 = plot(ensemble_nsum, title = "Neural SDE: Before Training")
scatter!(p1,t,sde_data',lw=3)
scatter(t,sde_data[1,:],label="data")
scatter!(t,Flux.data(pred[1,:]),label="prediction")
```
Now just as with the neural ODE we define a loss function:
```julia
function predict_n_sde()
n_sde(u0)
end
loss_n_sde1() = sum(abs2,sde_data .- predict_n_sde())
loss_n_sde10() = sum([sum(abs2,sde_data .- predict_n_sde()) for i in 1:10])
Flux.back!(loss_n_sde1())
data = Iterators.repeated((), 10)
opt = ADAM(0.025)
cb = function () #callback function to observe training
sample = predict_n_sde()
# loss against current data
display(sum(abs2,sde_data .- sample))
# plot current prediction against data
cur_pred = Flux.data(sample)
pl = scatter(t,sde_data[1,:],label="data")
scatter!(pl,t,cur_pred[1,:],label="prediction")
display(plot(pl))
end
# Display the SDE with the initial parameter values.
cb()
```
Here we made two loss functions: one which uses single runs of the SDE and another which
uses multiple runs. This is beceause an SDE is stochastic, so trying to fit the mean to
high precision may require a taking the mean of a few trajectories (the more trajectories
the more precise the calculation is). Thus to fit this, we first get in the general area
through single SDE trajectory backprops, and then hone in with the mean:
```julia
Flux.train!(loss_n_sde1 , ps, Iterators.repeated((), 100), opt, cb = cb)
Flux.train!(loss_n_sde10, ps, Iterators.repeated((), 20), opt, cb = cb)
```
And now we plot the solution to an ensemble of the trained neural SDE:
```julia
dudt_(u,p,t) = Flux.data(dudt(u))
g(u,p,t) = mp.*u
nprob = SDEProblem(dudt_,g,u0,(0.0f0,1.2f0),nothing)
ensemble_nprob = EnsembleProblem(nprob)
ensemble_nsol = solve(ensemble_nprob,SOSRI(),trajectories = 100)
ensemble_nsum = EnsembleSummary(ensemble_nsol)
p2 = plot(ensemble_nsum, title = "Neural SDE: After Training", xlabel="Time")
scatter!(p2,t,sde_data',lw=3,label=["x" "y" "z" "y"])
plot(p1,p2,layout=(2,1))
```

(note: for simplicity we have used a constant `mp` vector, though once can `param` and
train this value as well.)
Try this with GPUs as well!
### Neural Jump Diffusions (Neural Jump SDE) and Neural Partial Differential Equations (Neural PDEs)
For the sake of not having a never-ending documentation of every single combination of CPU/GPU with
every layer and every neural differential equation, we will end here. But you may want to consult
[this blog post](http://www.stochasticlifestyle.com/neural-jump-sdes-jump-diffusions-and-neural-pdes/) which
showcases defining neural jump diffusions and neural partial differential equations.
## A Note About Performance
DiffEqFlux.jl implements all interactions of automatic differentiation systems to satisfy completeness, but that
does not mean that every combination is a good combination.
### Performance tl;dr
- Use `diffeq_adjoint` with an out-of-place non-mutating function `f(u,p,t)` on ODEs without events.
- Use `diffeq_rd` with an out-of-place non-mutating function (`f(u,p,t)` on ODEs/SDEs, `f(du,u,p,t)` for DAEs,
`f(u,h,p,t)` for DDEs, and [consult the docs](http://docs.juliadiffeq.org/latest/index.html) for other equations)
for non-ODE neural differential equations or ODEs with events
- If the neural network is a sufficiently small (or non-existant) part of the differential equation, consider
`diffeq_fd` with the mutating form (`f(du,u,p,t)`).
- Always use GPUs if the majority of the time is in larger kernels (matrix multiplication, PDE convolutions, etc.)
### Extended Performance Discussion
The major options to keep in mind are:
- in-place vs out-of-place: for ODEs this amounts to `f(du,u,p,t)` mutating `du` vs `du = f(u,p,t)`. In almost all
scientific computing scenarios with floating point numbers, `f(du,u,p,t)` is highly preferred. This extends to
dual numbers and thus forward difference (`diffeq_fd`). However, reverse-mode automatic differentiation as implemented
by Flux.jl's Tracker.jl does not allow for mutation on its `TrackedArray` type, meaning that mutation is supported
by `Array{TrackedReal}`. This fallback is exceedingly slow due to the large trace that is created, and thus out-of-place
(`f(u,p,t)` for ODEs) is preferred in this case.
- For adjoints, this fact is complicated due to the choices in the `SensitivityAlg`. See
[the adjoint SensitivityAlg options for more details](http://docs.juliadiffeq.org/latest/analysis/sensitivity.html#Options-1).
When `autojacvec=true`, a backpropogation is performed by Tracker in the intermediate steps, meaning the rule about mutation
applies. However, the majority of the computation is not hte `v^T*J` computation of the backpropogation, so it is not always
obvious to determine the best option given that mutation is slow for backprop but is much faster for large ODEs with many
scalar operations. But the latter portion of that statement is the determiner: if there are sufficiently large operations
which are dominating the runtime, then the backpropogation can be made trivial by using mutation, and thus `f(u,p,t)` is
more efficient. One example which falls into this case is the neural ODE which has large matrix multiply operations. However,
if the neural network is a small portion of the equation and there is heavy reliance on directly specified nonlinear forms
in the differential equation, `f(du,u,p,t)` with the option `sense=SensitivityAlg(autojacvec=false)` may be preferred.
- `diffeq_adjoint` currently only applies to ODEs, though continued development will handle other equations in the future.
- `diffeq_adjoint` has O(1) memory with the default `backsolve`. However, it is known that this is unstable on many equations
with high enough stiffness (this is a fundamental fact of the numerics, see
[the blog post for details and an example](https://julialang.org/blog/2019/01/fluxdiffeq). Likewise, this instability is not
often seen when training a neural ODE against real data. Thus it is recommended to try with the default options first, and
then set `backsolve=false` if unstable gradients are found. When `backsolve=false` is set, this will trigger the `SensitivityAlg`
to use [checkpointed adjoints](http://docs.juliadiffeq.org/latest/analysis/sensitivity.html#Options-1), which are more stable
but take more computation.
- When the equation has small enough parameters, or they are not confined to large operations, `diffeq_fd` will be the fastest.
However, as it is well-known, forward-mode AD does not scale well for calculating the gradient with respect to large numbers
of parameters, and thus it will not scale well in cases like the neural ODE.
## API Documentation
### DiffEq Layer Functions
- `diffeq_rd(p,prob, args...;u0 = prob.u0, kwargs...)` uses Flux.jl's
reverse-mode AD through the differential equation solver with parameters `p`
and initial condition `u0`. The rest of the arguments are passed to the
differential equation solver. The return is the DESolution. Note: if you
use this function, it is much better to use the allocating out of place
form (`f(u,p,t)` for ODEs) than the in place form (`f(du,u,p,t)` for ODEs)!
- `diffeq_fd(p,reduction,n,prob,args...;u0 = prob.u0, kwargs...)` uses
ForwardDiff.jl's forward-mode AD through the differential equation solver
with parameters `p` and initial condition `u0`. `n` is the output size
where the return value is `reduction(sol)`. The rest of the arguments are
passed to the differential equation solver.
- `diffeq_adjoint(p,prob,args...;u0 = prob.u0, kwargs...)` uses adjoint
sensitivity analysis to "backprop the ODE solver" via DiffEqSensitivity.jl.
The return is the time series of the solution as an array solved with parameters
`p` and initial condition `u0`. The rest of the arguments are passed to the
differential equation solver or handled by the adjoint sensitivity algorithm
(for more details on sensitivity arguments, see
[the diffeq documentation](http://docs.juliadiffeq.org/latest/analysis/sensitivity.html#Adjoint-Sensitivity-Analysis-1)).
### Neural DE Layer Functions
- `neural_ode(model,x,tspan,args...;kwargs...)` defines a neural ODE layer where
`model` is a Flux.jl model, `x` is the initial condition, `tspan` is the
time span to integrate, and the rest of the arguments are passed to the ODE
solver. The parameters should be implicit in the `model`.
- `neural_dmsde(model,x,mp,tspan,args...;kwargs)` defines a neural multiplicative
SDE layer where `model` is a Flux.jl model, `x` is the initial condition,
`tspan` is the time span to integrate, and the rest of the arguments are
passed to the SDE solver. The noise is assumed to be diagonal multiplicative,
i.e. the Wiener term is `mp.*u.*dW` for some array of noise constants `mp`.

